9.9.2. Marking up Dataset page with Schema.org & Bioschemas for SEO¶

9.9.2.1. Main Objectives¶
The main purpose of this recipe is:
To embed
Schema.orgmarkup in a web page representing a dataset.
Actions.Objectives.Tasks |
Input |
Output |
|---|---|---|
Data Formats |
Terminologies |
Models |
|---|---|---|
9.9.2.2. Graphical Overview¶
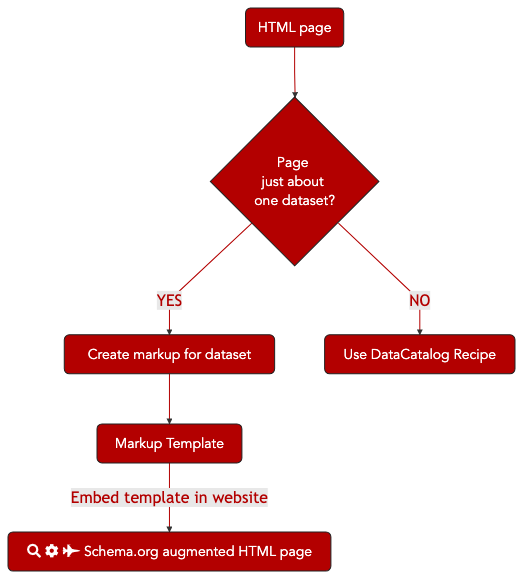
Fig. 9.6 The process of annotating a dataset webpage with bioschema markup for Search Engine discovery.¶
9.9.2.3. Method¶
We will outline the steps for marking up a page in your site that is about a specific dataset that you publish. The resulting markup will be compliant with both Google’s Dataset markup guidelines and the Bioschemas Dataset Profile. The resulting webpage will be indexable by the major search engines and should eventually appear in Google’s Dataset Search Tool.
We will use UniProtKB as an example for this recipe.
Identify the page in your site about a specific dataset, e.g. https://www.uniprot.org/uniprot/
Open the Bioschemas Generator
Select
Datasetfrom the Bioschemas Profile dropdownEnter the URL of the page in URL box, e.g.
https://www.uniprot.org/uniprot/Click on the
Show Formbutton
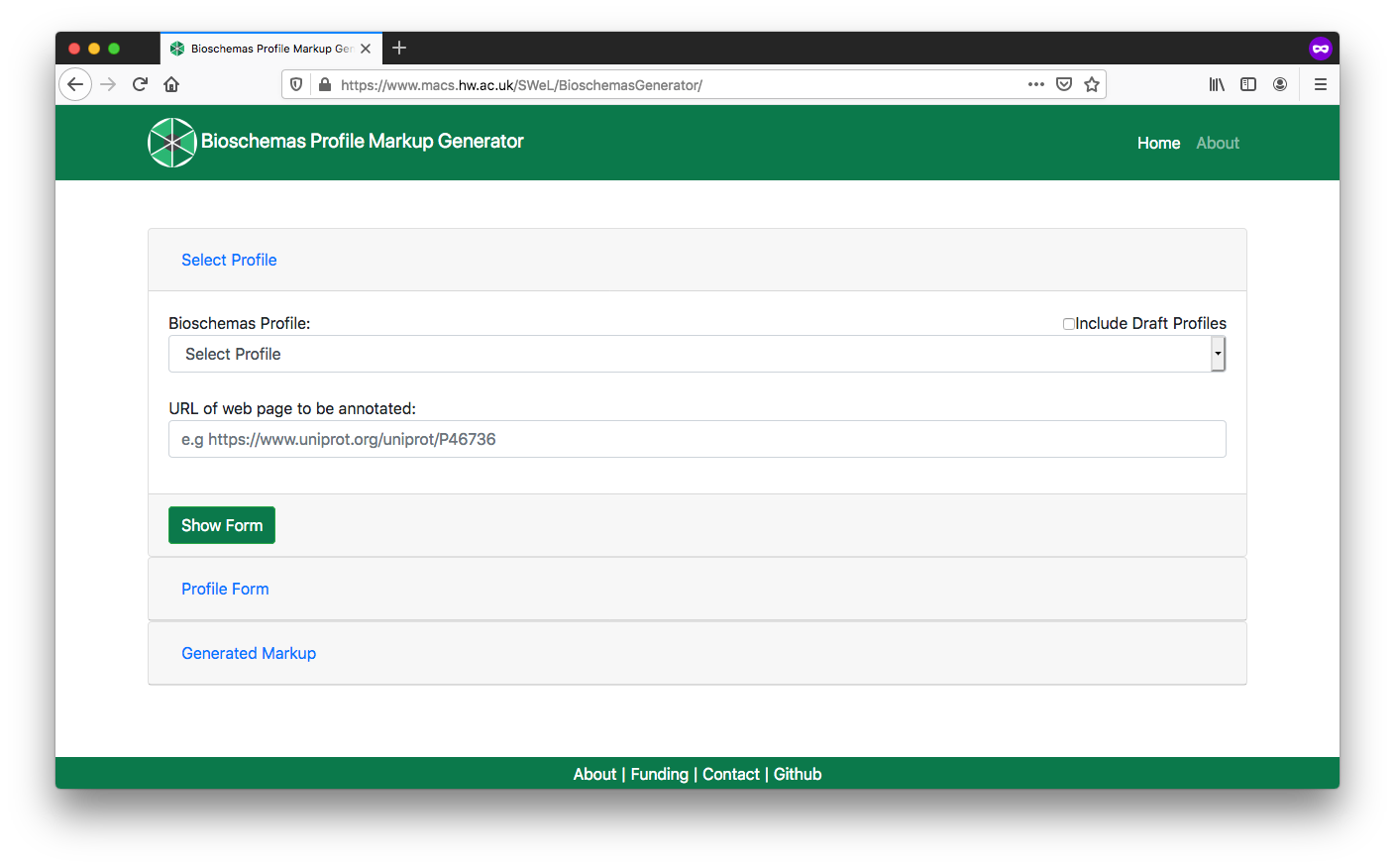
Fig. 9.7 Bioschemas Generator start screen.¶
Complete the profile form with the data relevant for your page. Once completed, click on the
Generate MarkupbuttonYou should complete all Minimum properties and as many Recommended properties as possible. You can show/hide properties using the
Additional Propertiesbuttons.Where possible you should link to other resources. The Bioschemas Generator does not make this as simple as it could, but you can do it in step 5 once you have generated your markup, e.g. our dataset will link to a page with DataCatalog markup in rather than repeating all the properties for now we will just enter a
urland no other propertiesThe form defaults to the data type with the first alphabetical character, e.g. for
identifier, this defaults toPropertyValuebutTextorURLwill be more appropriate in most casesThe right side of the screen gives examples for properties, where these have been provided by the Bioschemas profile authors. Click on the
Showbutton to see the example for a specific property. Click onMinimum,Recommended, orOptionalto expand/contract the section and see the properties contained at that marginality level
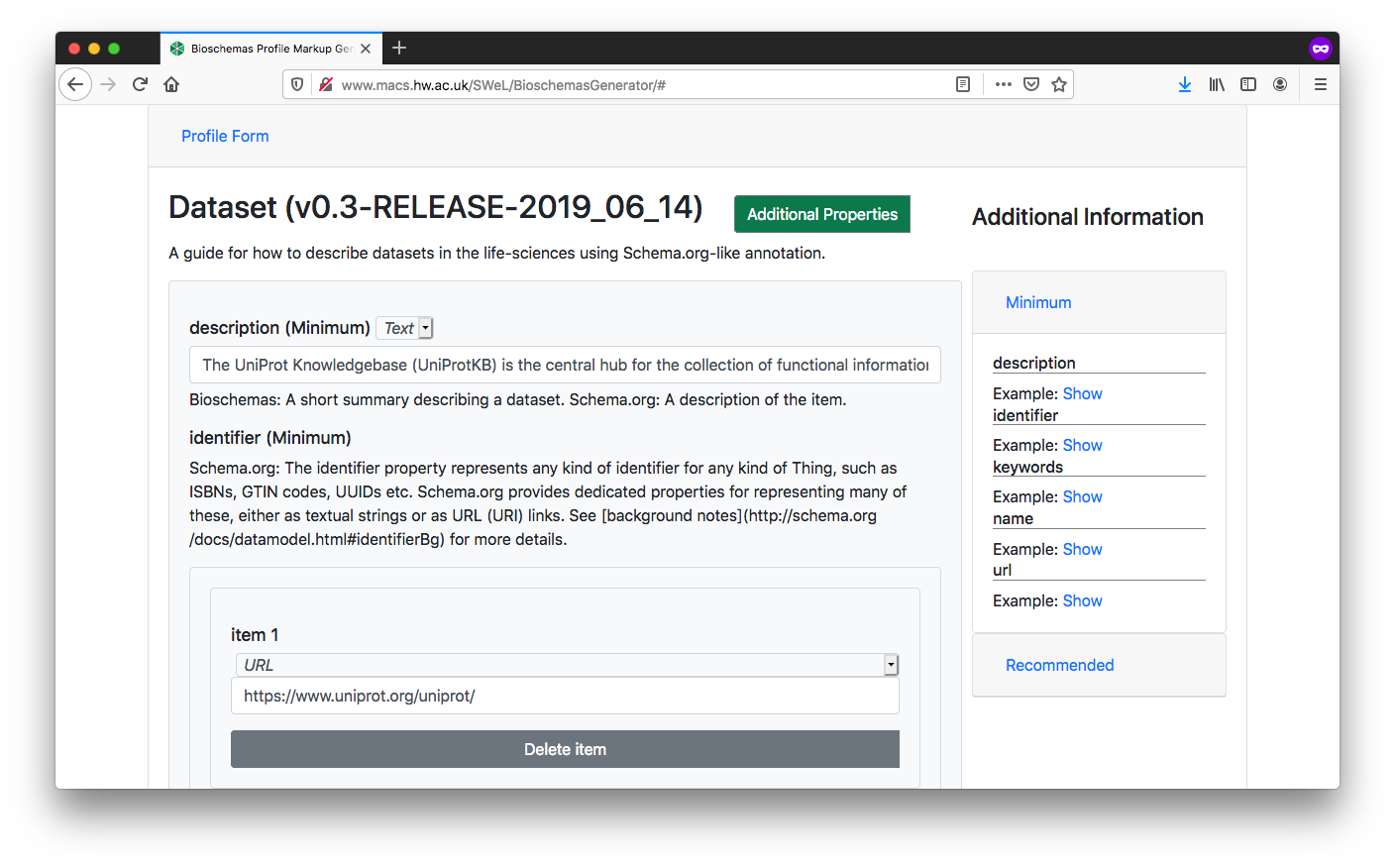
Fig. 9.8 Bioschemas Generator Dataset profile form.¶
You will now see the generated markup in
JSON-LDformat. You can click on theMicrodataandRDFatabs to see the same content rendered in the different formats. However, we recommend the use ofJSON-LD. For our UniProtKB example, we get the following markup<script type="application/ld+json" > { "@context": "https://schema.org", "@id": "https://www.uniprot.org/uniprot/", "@type": "Dataset", "citation": [ { "@id": "https://doi.org/10.1093/nar/gky1049", "@type": "CreativeWork" } ], "creator": [ { "@context": "https://schema.org", "@type": "Organization", "dct:conformsTo": "https://bioschemas.org/profiles/Organization/0.2-DRAFT-2019_07_19", "description": "The mission of UniProt is to provide the scientific community with a comprehensive, high quality and freely accessible resource of protein sequence and functional information. ", "name": "UniProt Consortium" } ], "dct:conformsTo": "https://bioschemas.org/profiles/Dataset/0.3-RELEASE-2019_06_14", "description": "The UniProt Knowledgebase (UniProtKB) is the central hub for the collection of functional information on proteins, with accurate, consistent and rich annotation. In addition to capturing the core data mandatory for each UniProtKB entry (mainly, the amino acid sequence, protein name or description, taxonomic data and citation information), as much annotation information as possible is added.", "distribution": { "@id": "https://www.uniprot.org/downloads#uniprotkblink", "@type": "DataDownload" }, "identifier": [ "https://www.uniprot.org/uniprot/" ], "includedInDataCatalog": [ { "@context": "https://schema.org", "@type": "DataCatalog", "dct:conformsTo": "https://bioschemas.org/profiles/DataCatalog/0.3-RELEASE-2019_07_01", "description": "", "keywords": [], "name": "", "url": "https://uniprot.org" } ], "keywords": [ "Protein", "Protein annotation" ], "license": "https://creativecommons.org/licenses/by/4.0/", "name": "UniProtKB", "url": "https://www.uniprot.org/uniprot/" } </script >
Download or copy and paste the generated markup
Make adjustments for any bits that could not be properly entered through the form.
For example, for our generated markup we would change the
includedInDataCatalogso that it provides a direct link rather than repeating the properties. We would replace"includedInDataCatalog": [ { "@context": "https://schema.org", "@type": "DataCatalog", "dct:conformsTo": "https://bioschemas.org/profiles/DataCatalog/0.3-RELEASE-2019_07_01", "description": "", "keywords": [], "name": "", "url": "https://uniprot.org" } ],
with
"includedInDataCatalog": { "@type": "DataCatalog", "@id": "https://uniprot.org" },
You can test that your JSON-LD is valid syntax, and visualise your markup using the JSON-LD Playground.
Once you are happy with your markup, include the
JSON-LD, script tags and all, at the bottom of your HTML page template.Make sure that this is before the closing
</html>tagIf you have multiple datasets released through your site, then you should make a template for your datasets. In your template you should replace the values in your markup that will change from dataset to dataset with variables. Your web page templating system will replace the variables with values from your database. For example, the follow snippet uses variables of the form
%%%PAGEURL%%%<script type="application/ld+json"> { "@context": "https://schema.org", "@id": "%%%PAGEURL%%%", "@type": "Dataset", "citation": [ { "@id": "%%%DOI%%%", "@type": "CreativeWork" } ], "creator": [ { "@context": "https://schema.org", "@type": "Organization", "dct:conformsTo": "https://bioschemas.org/profiles/Organization/0.2-DRAFT-2019_07_19", "description": "The mission of UniProt is to provide the scientific community with a comprehensive, high quality and freely accessible resource of protein sequence and functional information. ", "name": "UniProt Consortium" } ... ] } </script>
Your site should now generate dataset pages with embedded markup.
Once you have deployed this on your web server, you can test it with the Bioschemas Validator which scrapes the markup from your page and allows you to test it against various Bioschemas profiles1.
9.9.2.5. References¶
References
1: The Bioschemas Validator is currently in an early alpha release and does not include all the profiles.
9.9.2.6. Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
Heriot Watt University |
Writing - Original Draft |
||||
ZB MED Information Centre for life sciences |
Writing - Review & Editing |
||||
University of Oxford |
Writing - Review & Editing |


