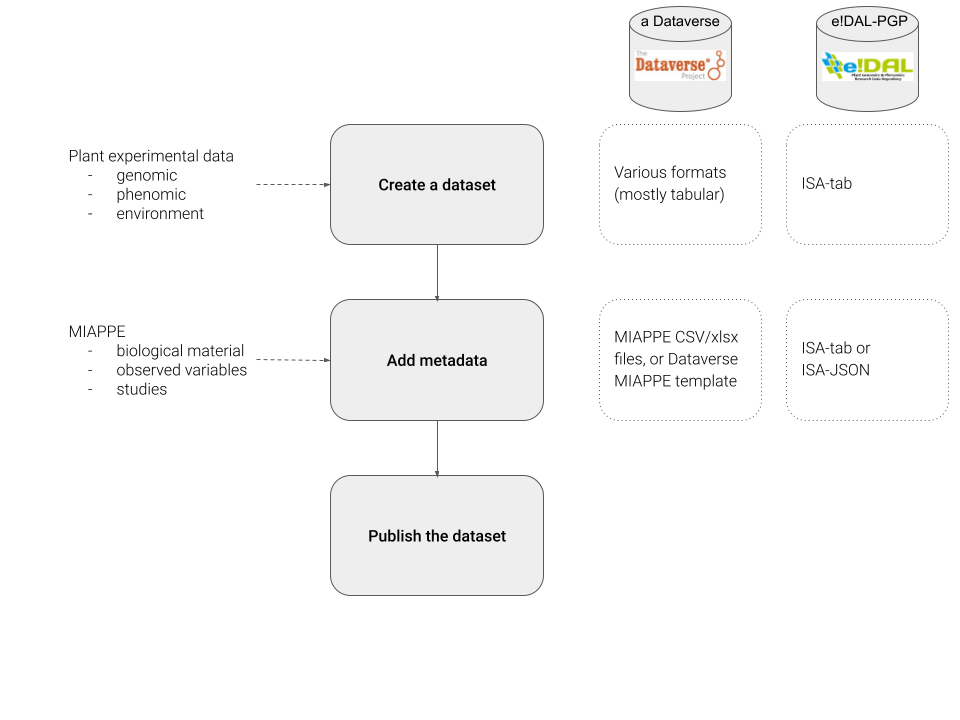
Publication of plant experimental data in generic data repositories¶

Main Objective¶
Generic data repositories offer high level metadata, but they are not sufficient to guarantee interoperability and the possibility to reuse your data later. They must be completed by the Minimum Information About Plant Phenotyping Experiments (MIAPPE) international standard. The Plant Sciences domain page of the RDMkit describes how MIAPPE is needed and used to clearly document the biological material used as well as the traits, environmental and phenotypical, recorded and computed. Those MIAPPE metadata can be stored using: MIAPPE full template, selected sheets from the full template (recommended since it can be stored as CSV), and ISA Tab archives. The first is the easiest to use, while the latter require a certain level of automation and toolsets.
The main objective of the recipe is to guide plant scientists during the submission of experimental data to generic repositories such as Dataverses, e!Dal-PGP, or Zenodo. This includes:
Dataset creation
Addition of mandatory metadata to the dataset
Publication of the dataset.
Summary¶
This recipe describes best practices for submitting plant experimentation data to generic data repositories (e.g. e!DAL-PGP, Dataverses such as recherche.data.gouv, dmportal.biodata.pt and Jülich DATA). This will allow data reuse according to FAIR principles, and especially :
Ensure visibility and reuse of genetic and phenomic datasets via minimal and sufficient description: data type, organism, list of plant material used, experimental metadata including methods and protocols, etc.
Maximize data visibility by allowing their indexation in international portals
Ensure the interoperability of data sets in relation to a coherent identification of plant material used in experiments of various kinds (phenotyping, genotyping, genomics, etc.)
FAIRification Objectives, Inputs and Outputs¶
Actions.Objectives.Tasks |
Input |
Output |
|---|---|---|
Step-by-Step process for data submission and publication in Dataverses¶
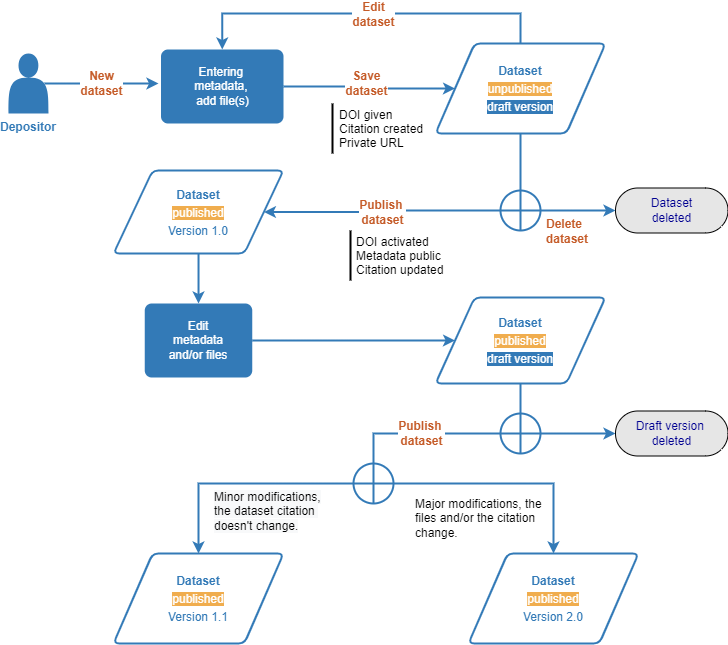
Example of global dataset management scheme for dataverses
Step 1: Dataset creation¶
Find an appropriate dataverse¶
You need first to select the appropriate dataverse and/or sub-dataverse for your use case depending on the constraints of your consortium or institute. The guidelines explained below are applicable to all of those dataverse instance. You will find below a non -exhaustive list of dataverse instances. Also, find the right sub-dataverse for your submission, like the one of the research group or the project you belong to: dataverses can contain dataverses.
-
Open to submission from any consortium involving at least one member of a french research institution.
Examples: Data INRAE and CIRAD.
Jülich DATA (NRW - DE)
Open to submission from any research activity done from partners at Forschungszentrum Jülich.
Meant for data and software submissions.
Maintained by the central library of FZJ.
Examples under subject “Agricultural Sciences”.
dmportal.biodata.pt (PT)
Open to submission of biological data from Portuguese research & innovation institutions.
Example: Plant BioDataVerse.
Authenticate¶
Click on “Log in” in the top right corner, and choose the right authentication mechanism. Use your institutional login if it is present:
in recherche.data.gouv: LDAP of your institute (only french institutes available)
in Jülich DATA: Helmholtz AAI (federated, non-german institutes are also available)
in dmportal.biodata.pt: registration form.
Connecting with an ORCID, Github account, or Google login can also be available, depending on the dataverse instance.
Create a new dataset¶
Create a new dataset
Fig. 30 Create a new dataset in a dataverse¶
If you have permissions issues, ask for support from the dataverse owner (data.inrae: urgi-data@inrae.fr ; Jülich DATA: ask for support from central library team (forschungsdaten@fz-juelich.de or via chat. You might need account validation or specific permissions to create datasets.
Fill in the metadata¶
Fill in the metadata
Fig. 31 Fill in the metadata of the dataset¶
For a generic template, the required fields of this form are Title, Contact, Author, Description, Subject, Kind of Data. Metadata Tip: After adding the dataset, click the Edit Dataset button to add more metadata.
Save the dataset¶
The DOI will be automatically generated but the dataset will remain in draft mode (with the “Draft” and “Unpublished” tags) until it is voluntarily published.
Manage collaborative editing and access rights¶
It is possible to ask partners to complete certain metadata by giving them the appropriate rights.
Ask the partner to create an account on the Dataverse containing the dataset, and to give you its username.
On the dataset, click on Edit > Permissions > Dataset.
Click on the Assign Roles to Users/Groups button.
Fill in the name of the producer and assign the role of Contributor
Save changes
Step 2: Add mandatory metadata for plant phenotyping data¶
Note
This section is also applicable to Zenodo.
Note
Some dataverse instances (such as dmportal.biodata.pt) have a MIAPPE metadata section that allows the use of dataverse web pages to store Biological material, Observation Variable and Study metadata. In that case you can use your dataverse’s MIAPPE metadata instead of the MIAPPE metadata files described in this step.
The description of the plant material used in the experimentation proposed by MIAPPE is common to all types of experimentation (phenotypic, genetic, omic, etc…) and enables long-term reuse and interoperability between data sets. You can either use the full MIAPPE template or only selected sheets, available as individual file templates which can be found below.
Biological material¶
Use BiologicalMaterial.xlsx. This spreadsheet contains the following fields: Mandatory fields:
“Biological material ID” (ex: INRA:W95115_inra_2001): Lot number or material identifier in the data files
“Material source ID” (ex: INRA:B73_usda) OR “Accession_number” (B73_usda) + “Holding_institute” (ex: INRA)
Accession Number
Genus
Species Optional fields:
“Material source DOI”: accession DOI
Organism: NCBITAXON:4577
“Infraspecific name”: variety names, cultivar names, etc…
Genealogy:
Parent1or2_AccessionNumber
Parent1or2_TaxonGroup
Parent1or2_HoldingInstitutionName
Parent1or2_Type (father/mother/undefined)
All MIAPPE Biological Material fields (DM-40 to DM-56)
Free input: synonyms, project IDs, any relevant information on the plant material.
Observed variables¶
Use ObservedVariables.xlsx. This file is needed for the description of phenotyping experiments traits and methods.
Studies or experiments¶
It is recommended to list the experimentation done in this dataset, including in particular the GPS location, the site name and the environmental parameters which characterize the experimental sites. Use Studies.xlsx
How to add metadata files in a Dataverse¶
Click on “+ Upload Files” in the “Files” tab
Add the file(s). Please note that :
the file size is limited to 15.0 GB
compressed files are automatically decompressed at the time of import
tabbed files must use the “,” separator and “UTF-8” encoding to avoid problems during import (see the dedicated section in the user guide)
Fill in the “Description” field for each added file
Update the file labels by selecting “File options” > “Tags” for each file
Update the file labels
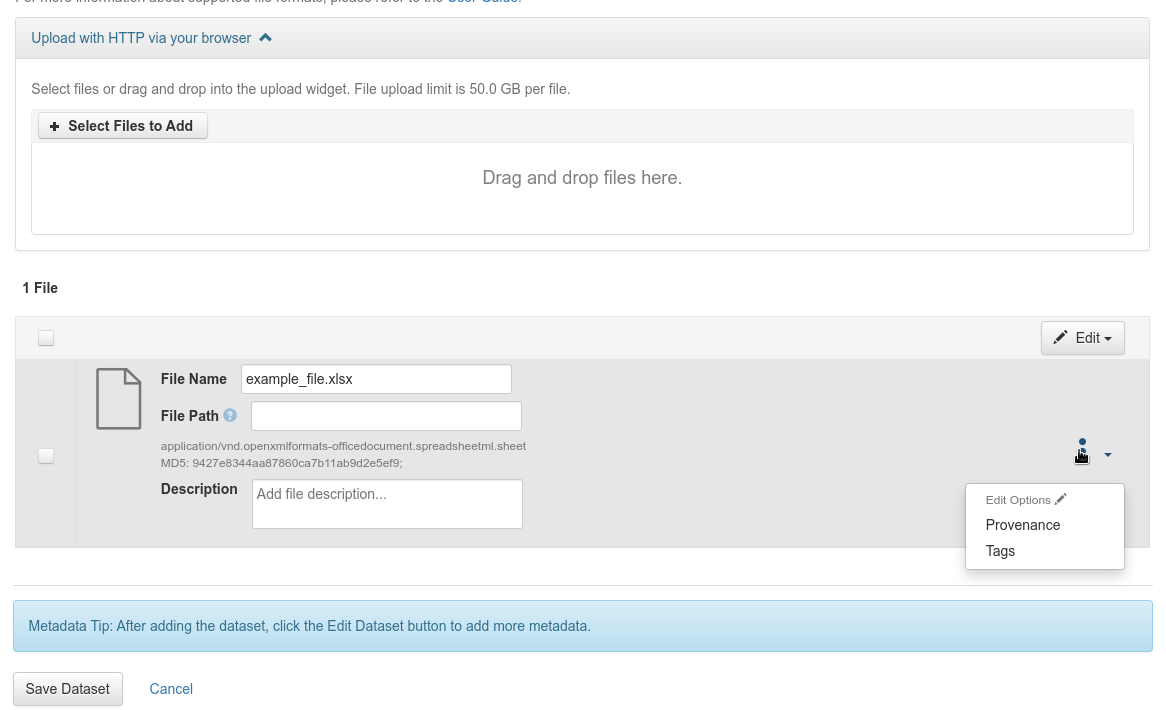
Fig. 32 Update the file labels¶
Add a custom label, “Biological_Material” or “Observed_Variable” depending on the file type. If the label exists, it will be available in the “File labels” section, otherwise you will have to create it in the “Customize file label” section and apply it.
Add a custom label
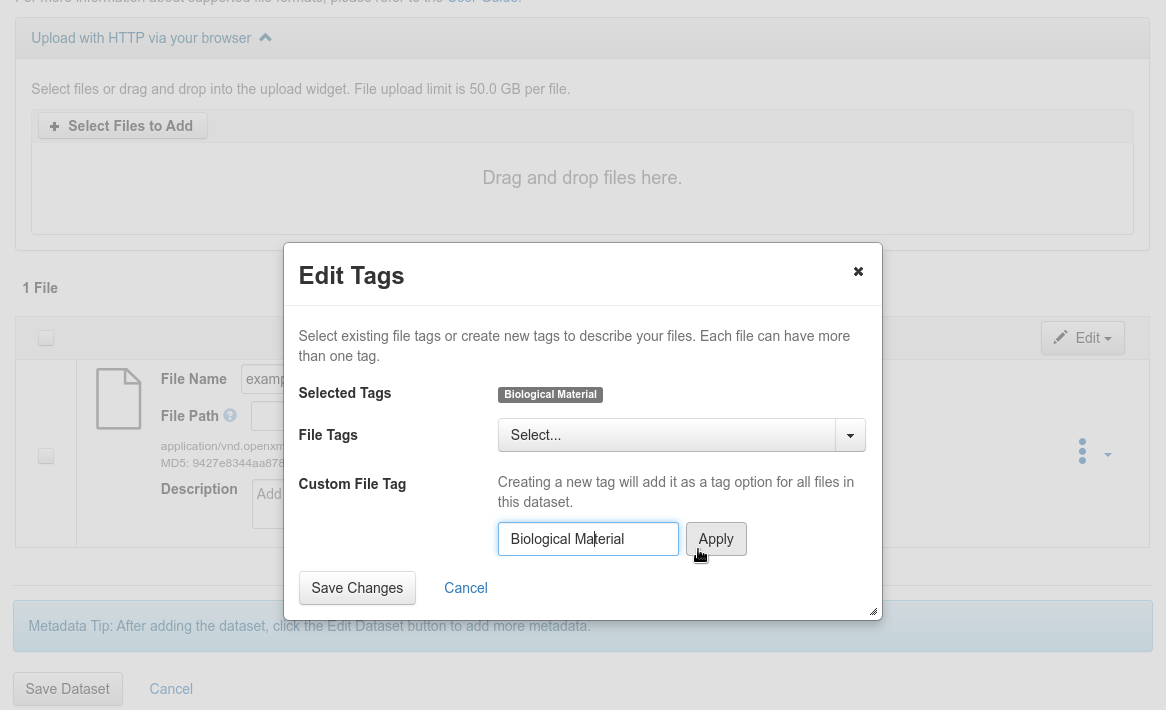
Fig. 33 Add a custom label¶
Save your modifications
Step 3: Add generic metadata to the dataset¶
These are the metadata that are not specific to plant sciences. Select the “Metadata” tab then click on the “add + edit metadata” button
Add and edit generic metadata
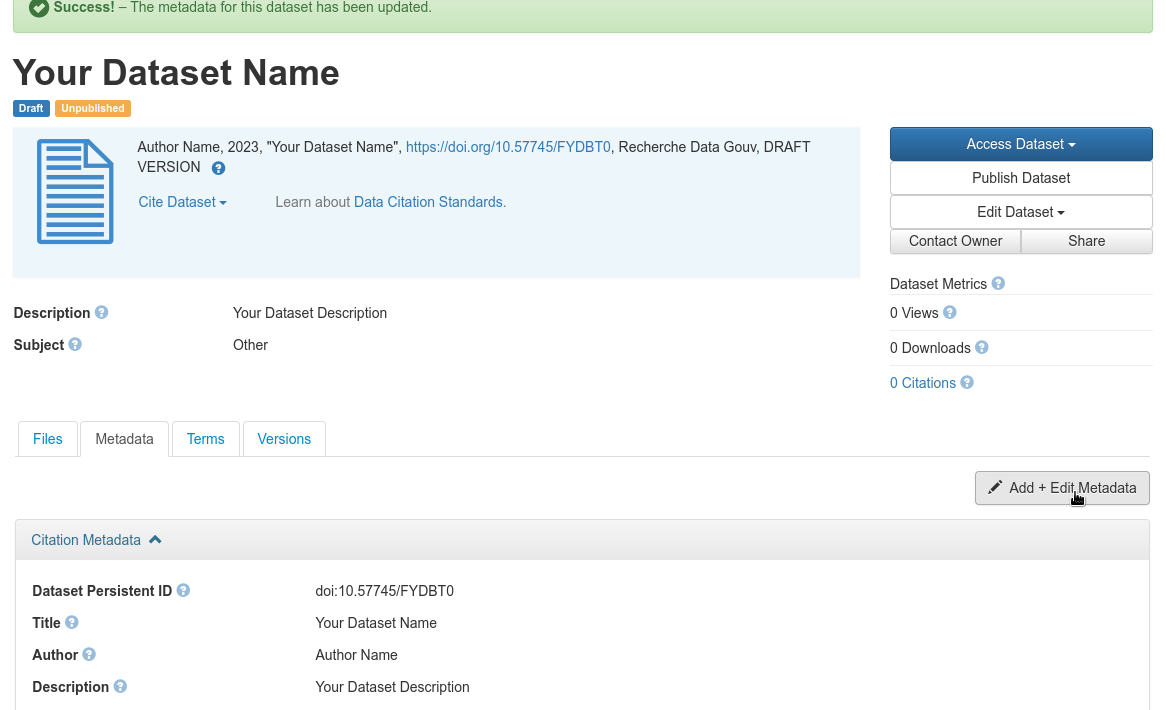
Fig. 34 Add and edit generic metadata¶
Mandatory metadata:
Language: in section “Citation Metadata” > “Language” : dataset language
Organism: in section “life science metadata” > “Organism” : species name
Recommended metadata:
Contributor
Keywords: In the list of Dataverses, Datasets and Files of your Dataverse, looking at existing entries for “Keyword Term” can help you find appropriate keywords.
Related publication
Grant Information
Project Information
Time Period Covered (start and end date)
Geospatial Metadata > Geographic Coverage
Step 4: Publish the dataset¶
Private publication: temporary¶
Give access to an unpublished dataset (private URL): Edit dataset > Private URL
Give access to an unpublished dataset
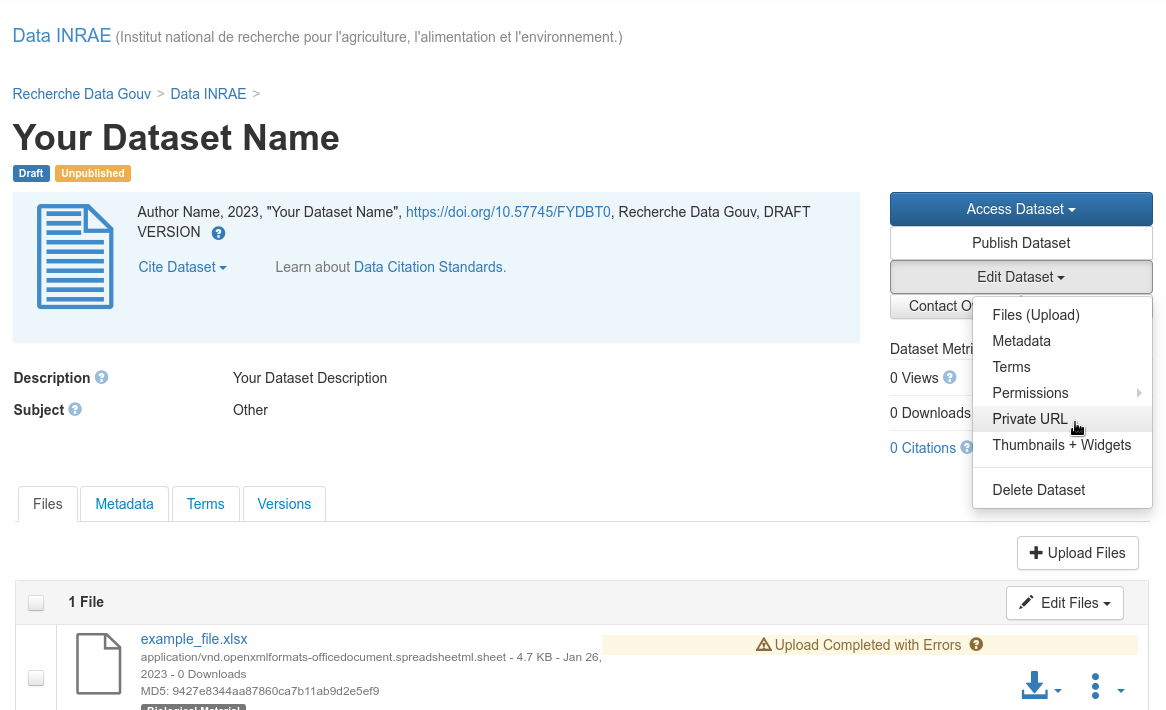
Fig. 35 Give access to an unpublished dataset¶
The submitter of a dataset can generate a private URL, to give access to a dataset not yet published to a person who has no access rights on it.
Create a private URL
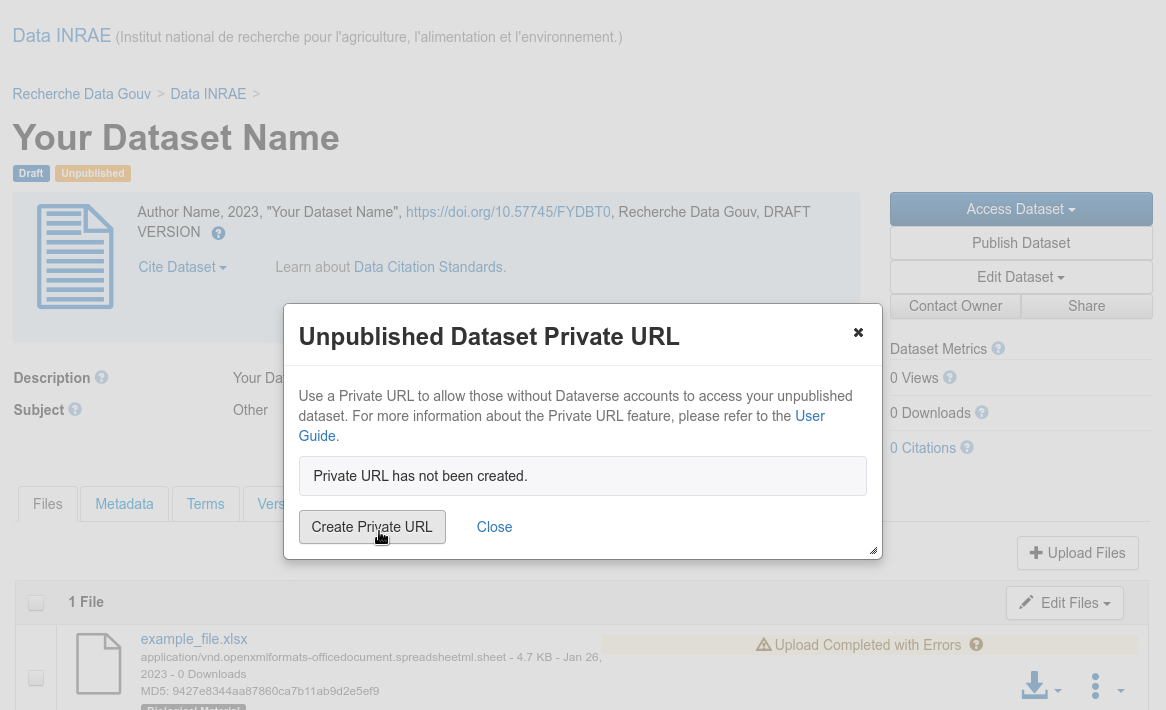
Fig. 36 Create a private URL¶
Unpublished dataset private URL
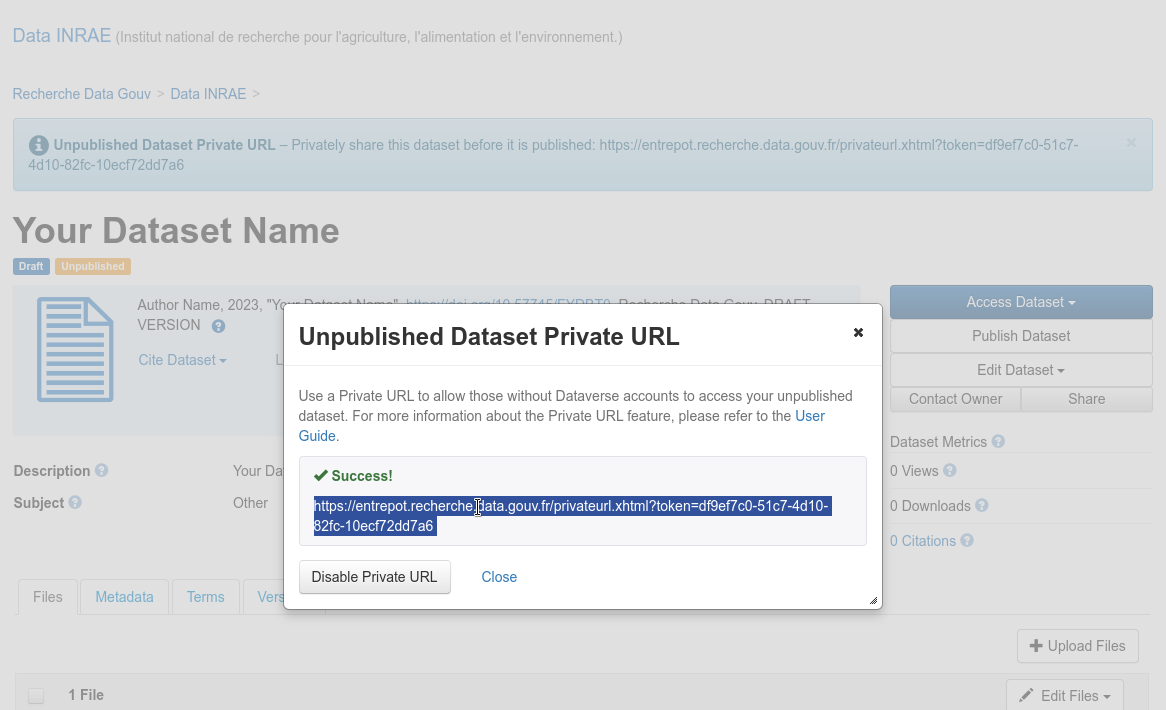
Fig. 37 Unpublished dataset private URL¶
Publication: definitive, with unlimited updates possible¶
Publish the dataset:
Publish the dataset
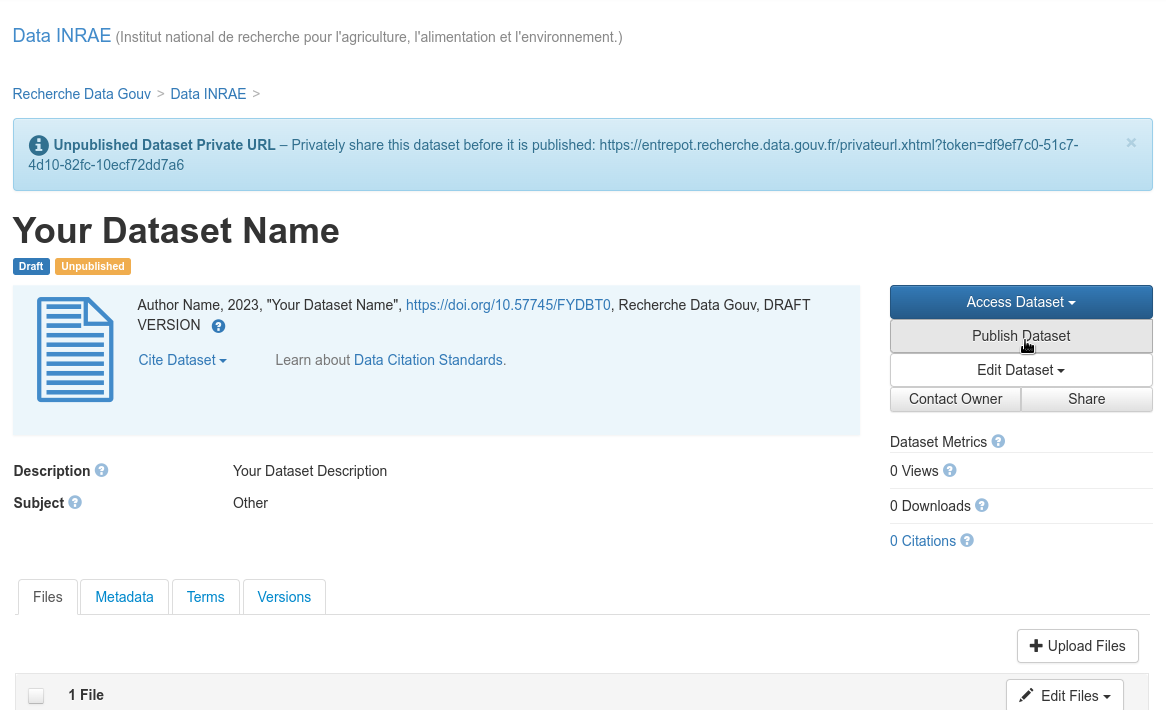
Fig. 38 Publish the dataset¶
Add an existing dataset to a specific dataverse¶
Open your dataset page and login, then choose “Link Dataset”:
Add an existing dataset to a specific dataverse
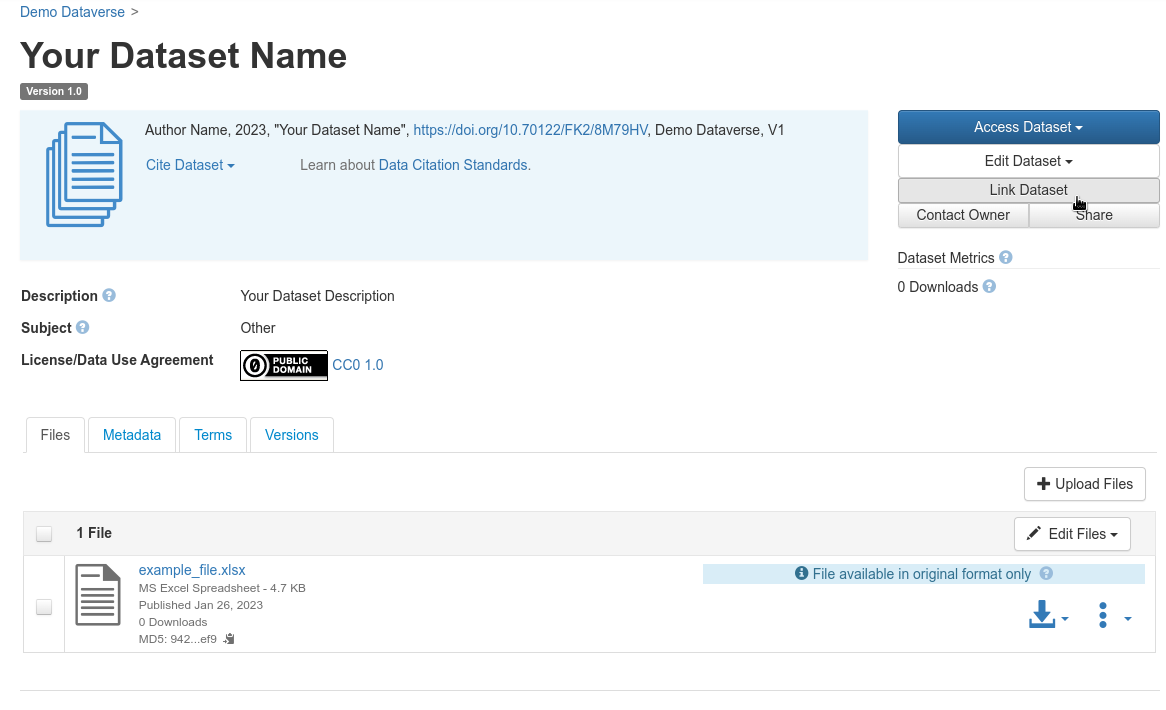
Fig. 39 Add an existing dataset to a specific dataverse¶
Find the dataverse in which you want to add your dataset, select it and click on “Save Linked Dataset”.
Save Linked dataset
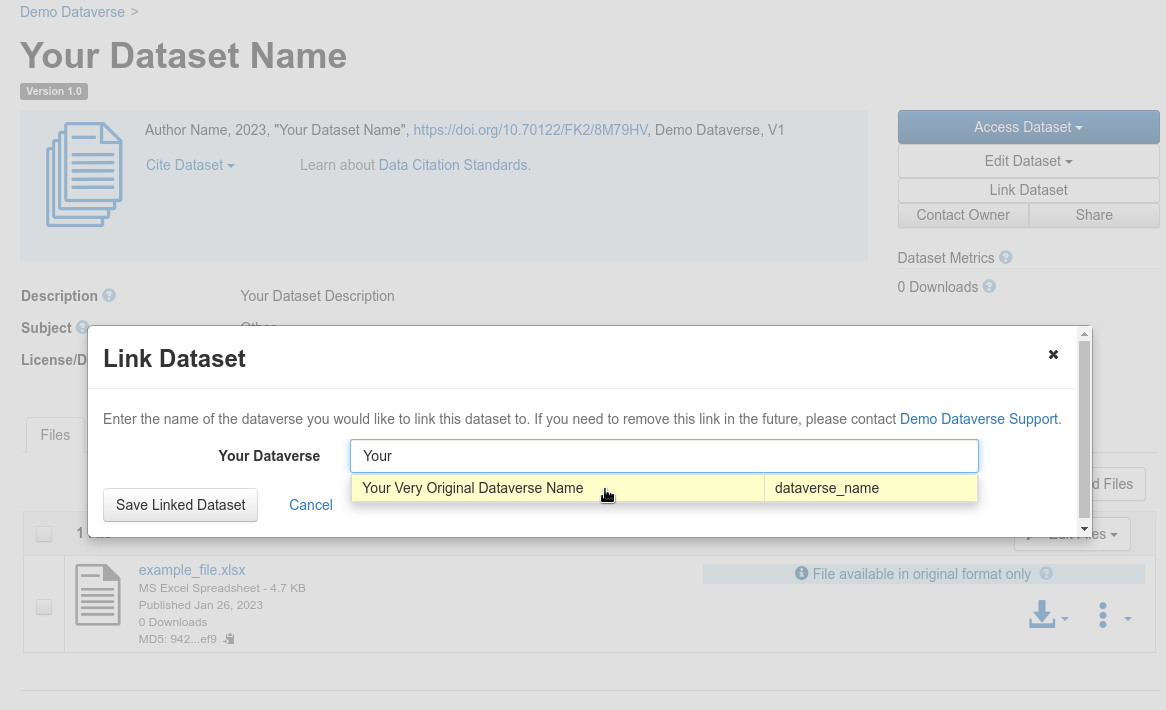
Fig. 40 Save linked dataset¶
Step-by-Step process for data submission and publication in e!DAL-PGP¶
Step 1: Dataset creation¶
Major aim of the eDAL-PGP repository is FAIR sharing plant related research data, which do not fit into domain-specific repositories and databases due to scope or volume. All available datasets are FAIR compliant and can be referenced via a persistent DOI. The submission procedure is open for all ELIXIR associated users over the LS AAI. The key feature of e!DAL-PGP is its user-friendly and simple data submission and internal review procedure
Authentication¶
The eDAL-PGP repository currently provides a local desktop application (Win/Unix/Mac) as well as a simple web-based submission tool to upload research data and initiate the intern review procedure. Both can be downloaded or rather accessed on the main project website.
The PGP repository
Fig. 41 The PGP repository¶
To authenticate for the submission process the LS AAI (formerly known as ELIXIR AAI) is used. Every user can select the identity provider of his home organization. If the home organization is not connected and therefore not in the selection list, the login is alternatively possible by using a third party provider like ORCID or Google. After successful login it is necessary to read the “Deposition and License Agreement” once.
Submit a new Dataset¶
For submitting a new dataset some technical metadata are necessary to describe the data and provide the opportunity for assigning a DOI later. Therefore the submission client provides a simple form-based user interface guiding the user through the different attributes and gives feedback in case of missing attributes. Some of the fields like “Description” are simple free text fields, while others like “Authors” provide more options when clicking into the field like the linkage to the ORCID registry to select the ORCID for every author.
Submit a new dataset
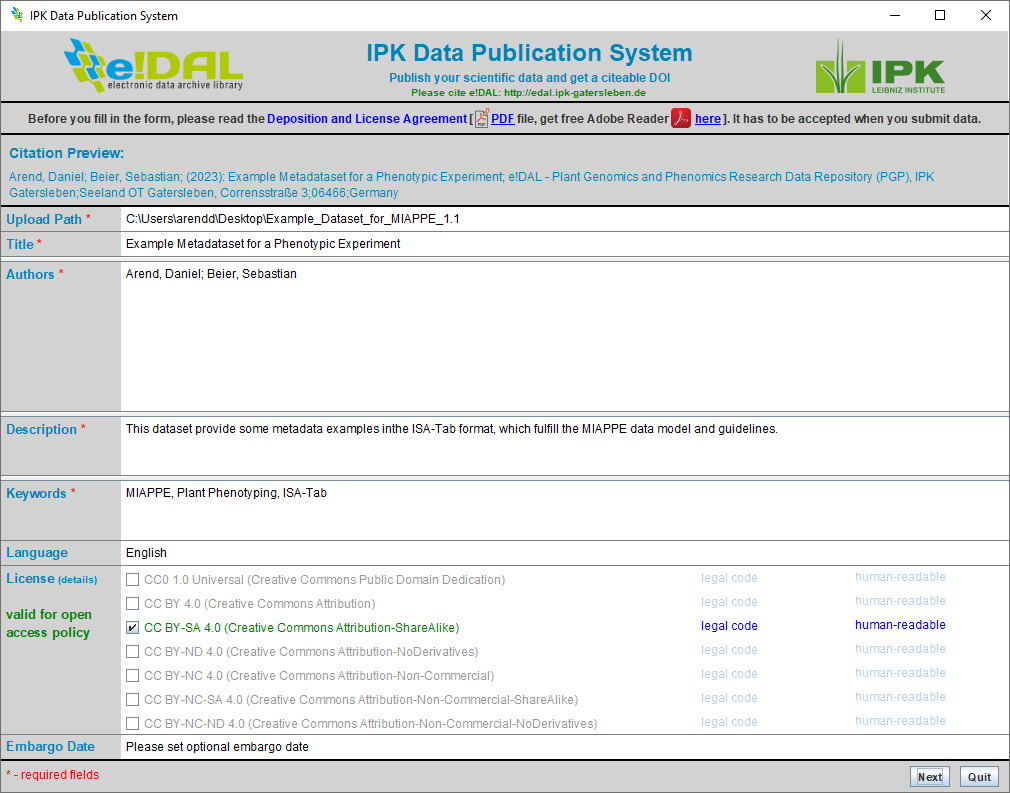
Fig. 42 Submit a new dataset¶
A dataset can contain single files as well as comprehensive folder structure. Compressed files like *.zip should be avoided, because this would prevent a later navigation on the content page for the published datasets. After entering all necessary metadata, it is necessary to accept the “Deposition and License Agreement” to start the submission process.
Start the submission process
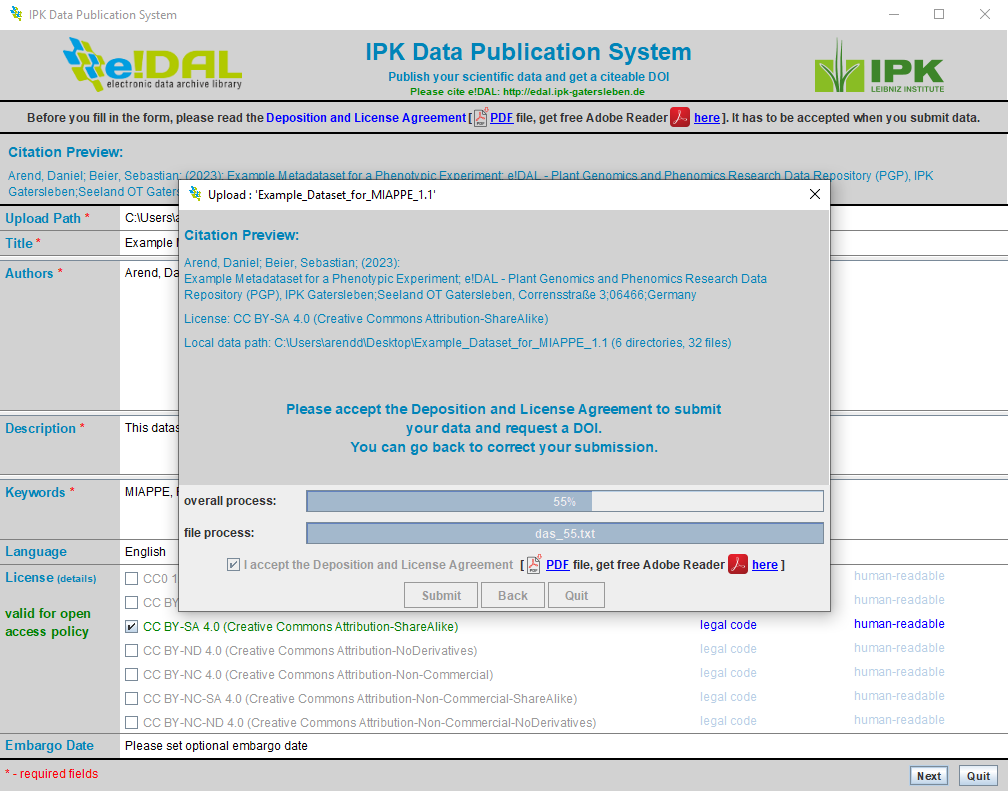
Fig. 43 Start the submission process¶
After finishing the upload process the submission is ready for the integrated review process. The data provider will receive an email to confirm, that his submission was successful and that the internal review war initiated.The reviewers will also automatically receive an email with a link providing a restricted access to the given dataset. After successfully checking some general requirement, such as a complete metadata or the use of open formats the data provider will receive an additional mail with the opportunity to assign the final DOI to publish and reference his datasets or alternatively cancel the submission. Usually there is an additional review necessary, because the data belongs to a corresponding research publication. Therefore the last email also contains a temporary preview link which the data provider can use to share the data with colleagues or additional reviewers and decide later if and when he wants to assign the final and immutable DOI.
Step 2: Add mandatory metadata for plant phenotyping data¶
Due to the generic concept of the underlying e!DAL infrastructure software the e!DAL-PGP repository requires only a standardized set of technical metadata inspired by the DublinCore metadata schema as well as the derived DataCite schema, which is needed to assign DOIs. Because of the scope of e!DAL-PGP on sharing mainly plant genotypic and phenotypic datasets the internal review focuses on providing corresponding semantic metadata and domain-specific. To add these metadata files, the data provider can directly add them using a suitable format, such as ISA-Tab or ISA-JSON for MIAPPE-compliant metadata.
Use the metadata folder to integrate metadata to the dataset
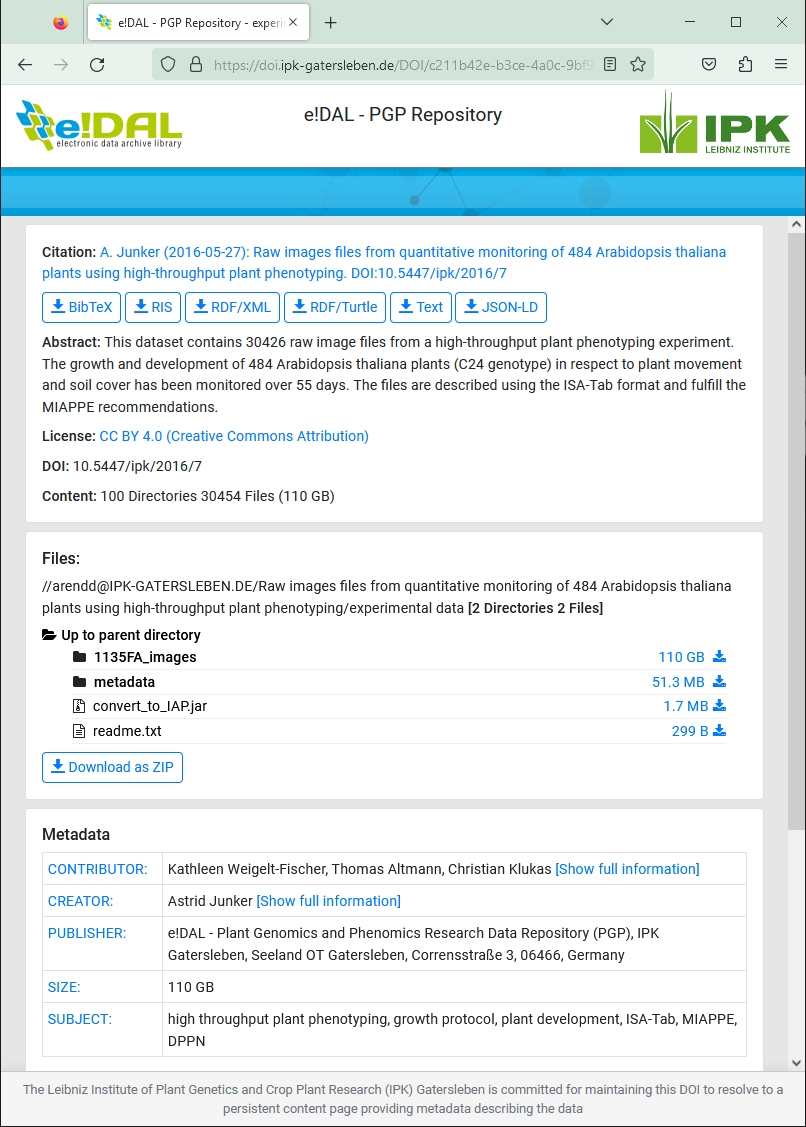
Fig. 44 Landing page for a phenotypic dataset in e!DAL-PGP¶
Step 3: Publish the dataset¶
Every uploaded dataset will get at the end of the submission and review process a DOI, which resolves to a landing page providing access to the datasets and the corresponding metadata. This DOI is immutable and can be used to reference and cite the datasets persistently. Before the final publication every data provider will receive a temporary preview link and decide on his own if and when he makes his datasets public. The preview link allows sharing the data before for additional review and communication with colleagues, but it is not intended to be used for permanent sharing. Additionally the mail contains two links to assign the final DOI or cancel the submission in case of any necessary changes.
Final approval email for data provider
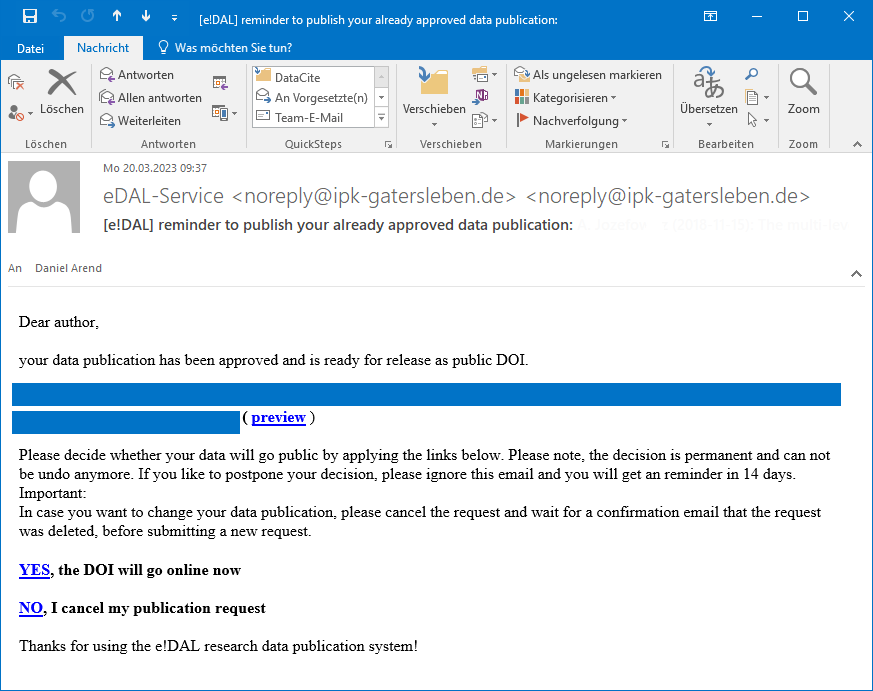
Fig. 45 Final approval email¶
Conclusion¶
In this recipe we have seen how to publish plant experimental data to generic data repositories in a MIAPPE-compliant way.
What to read next?¶
Documentation specific to:
the command-line tool DV Uploader:
Recherche Data Gouv:
Jülich DATA:
e!DAL-PGP:

FAIRsharing records appearing in this recipe:
- Comma-separated Values (CSV)
- DataCite Repository (DataCite)
- Digital Object Identifier (DOI)
- GitHub
- Investigation Study Assay Tabular (ISA-Tab)
- JavaScript Object Notation (JSON)
- Minimum Information about Plant Phenotyping Experiment (MIAPPE)
- NCBI Taxonomy (NCBITAXON)
- Plant Genomics and Phenomics Research Data Repository (PGP)
- Recherche Data Gouv
- The FAIR Principles (FAIR)
- Uniform Resource Locator (URL)
- Zenodo
Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
Université Paris-Saclay, INRAE, BioinfOmics |
Writing - Original Draft |
||||
Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK) |
Writing - Original Draft |
||||
Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) |
Writing - Original Draft |
||||
ITQB, NOVA University of Lisbon |
Writing - Original Draft |
||||
University of Lisbon |
Writing - Original Draft |
||||
Nicolas Francillonne |
Université Paris-Saclay, INRAE, BioinfOmics |
Writing - Original Draft |
|||
Université Paris-Saclay, INRAE, BioinfOmics |
Writing - Original Draft |
||||
Université Paris-Saclay, INRAE, BioinfOmics |
Writing - Original Draft |
||||
Université Paris-Saclay, INRAE, BioinfOmics |
Review & Editing |
||||
University of Oxford |
Review & Editing |