7. Ontology-related tools and services¶

7.1. Main Objectives¶
This recipe aims to provide an overview of tools available to perform a number of key operations using ontologies and relevant to FAIR processes: from ontology management to using ontology for annotation or performing ontology mapping.
It aims to serve as a starting point to identify tools for FAIRification tasks where ontologies and semantic frameworks are needed.
disclaimer
It is not intended to provide a comprehensive list covering all possible tools.
The lists of tools are generated either automatically by querying the bio.tools repository, or through manual curation. In this last instance, the list produced reflects what is being used in the industry and is influenced by the FAIRplus project partners that have been surveyed for the purpose of this work.
Warning
The content in these tables was generated in March 2021. For an updated contents, please check the FAIR tooling repository.
7.2. Requirements¶
recipe dependency:
knowledge requirement:
be familiar with ontologies and semantic annotation.
7.3. FAIRification Objectives, Inputs and Outputs¶
Actions.Objectives.Tasks |
Input |
Output |
|---|---|---|
7.4. Overview¶
The figure below shows different ontology-related operations and their relationships, together with related tools and recipes.
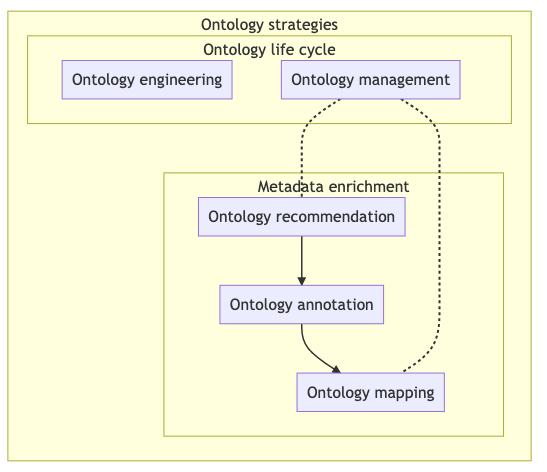
Fig. 7.2 Overview of key aspects in ontology associated processes¶
The table below is an overview of ontology strategies tools identified. Details of each tools are provided below.
Topic |
Curated tools |
Related tools in Bio.tools |
|---|---|---|
Ontology annotation |
ZOOMA |
bioBERT |
NCBI BioPortal Annotator |
PPR-SSM |
|
BioBert |
HPO2GO |
|
Termite |
Vapur |
|
PoolParty Semantic Suite |
matscholar |
|
OntoMaton |
CollaboNet |
|
Prodigy |
Calchas |
|
OntoText |
QTL TableMiner++(QTM) |
|
thbp |
||
Ontology mapping |
OxO |
meshr |
locdb |
||
Ontology management |
AberOWL |
ngly1 |
BioPortal |
Doc2Hpo |
|
Centree Ontology Manager |
PlanGexQ |
|
OLS |
GOcats |
|
Ontobee |
RDFScape |
|
PoolParty |
OntoBrowser |
|
QuickGO |
||
Circular Gene Ontology (CirGO) |
||
Ontology engineering |
eNanoMapper Slimmer |
|
OWLAPI |
||
Protégé |
||
ROBOT |
||
TopBraid Composer |
||
VocBench |
7.5. Operations¶
7.5.1. Ontology annotation¶
Ontoloy Annotation is the process of linking free text or data items to ‘tokens’ (defined terms from a lexicon) which provide semantic value. For example, “type 2 diabetes” can be annotated with corresponding term in the MONDO disease ontology.
Curated tools
Tool |
Description |
License |
Topics |
Resource Type |
How to use |
|---|---|---|---|---|---|
A tool for mapping free text annotations to ontology term based on a curated repository of annotation knowledge. |
Ontology and terminology, |
Web application, |
|||
Get annotations for biomedical text with classes from the ontologies. |
Ontology and terminology, |
Web application, |
|||
A biomedical language representation model designed for biomedical text mining tasks such as biomedical named entity recognition, relation extraction, question answering. |
text mining, |
Python |
|||
Semantic enrichment to unlock the value of unstructured text and simplify the identification of new potential biomarker leads from scientific text. |
Commercial license |
Ontology and terminology |
|||
Automate the handling of heterogeneous metadata systems and the creation of enterprise knowledge graphs.design knowledge graphs at your own pace and with speed. Create your own ontologies and custom schemes by reusing already existing ontologies such as FOAF, FIBO, schema.org and CHEBI, among others. Apply them to your existing taxonomies with ease. |
Commercial license |
Content enrichment, |
|||
A tool facilitating ontology search and tagging functionalities within Google Spreadsheets. |
Google Add-ons |
||||
A modern annotation tool for creating training and evaluation data for machine learning models. You can also use Prodigy to help you inspect and clean your data, do error analysis and develop rule-based systems to use in combination with your statistical models. |
Commercial license |
Data annotation |
Python, Web application,API |
||
Connect and publish complex enterprise knowledge with standard-compliant semantic graph database; Customize and apply analytics to link documents to graphs, extract new facts, classify and recommend content. |
Commercial license |
Related tools in Bio.Tools
Tool |
Description |
License |
Topics |
Resource Type |
|---|---|---|---|---|
A pre-trained weights of BioBERT, a language representation model for biomedical domain, especially designed for biomedical text mining tasks such as biomedical named entity recognition, relation extraction, question answering, etc. |
N/A |
Medicine, Ontology and terminology, Natural language processing |
Python |
|
Personalized PageRank and semantic similarity measures for linking entities found in documents to concepts from domain-specific ontologies. |
N/A |
Imaging, Natural language processing, Data mining, Genotype and phenotype, Ontology and terminology |
Java, Python |
|
Prediction of human phenotype ontology term associations using cross ontology annotation co-occurrences.Mapping between Human Phenotype Ontology (HPO) and Gene Ontology (GO) terms for the prediction of gene/protein - function - phenotype - disease associations. |
Pathology, Protein interactions, Genotype and phenotype, Ontology and terminology, Gene expression |
Command-line tool |
||
A Search Engine to Find Related Protein.Vapur is an online entity-oriented search engine for the COVID-19 anthology. Vapur is empowered with a semantic inverted index that is created through named entity recognition and relation extraction on CORD-19 abstracts. |
N/A |
Pathology, Ontology and terminology, Natural language processing, Enzymes |
Python |
|
A Python library for materials-focused natural language processing (NLP). Named Entity Recognition and Normalization Applied to Large-Scale Information Extraction from the Materials Science Literature. |
Chemistry, Ontology and terminology, Natural language processing |
Command-line tool |
||
Collaboration of deep neural networks for biomedical named entity recognition. |
Ontology and terminology, Natural language processing, Machine learning |
Command-line tool |
||
A web based framework that takes advantage of domain specific ontologies, and Natural Language Processing, aiming to empower exploration of biomedical resources via semantic-based querying and search. The NLP engine analyzes the input free-text query and translates it into targeted queries with terms from the underlying ontology. |
N/A |
Medical informatics, Ontology and terminology, Natural language processing, Bioinformatics |
Web application |
|
It is a command-line tool to retrieve and semantically annotate results obtained from QTL mapping experiments. It takes full-text articles from the Europe PMC repository as input and outputs the extracted QTLs into a relational database (SQLite) and text file (CSV). |
Ontology and terminology |
Command-line tool |
||
Mapping anatomical related entities to human body parts based on wikipedia in discharge summaries. |
N/A |
Anatomy, Ontology and terminology, Natural language processing |
7.5.2. Ontology mapping¶
The process of determining correspondences between equivalent concepts in alternative ontologies, and other vocabularies. This may include mapping to convey different levels of granularity.
Curated tools
Tool |
Description |
License |
Topics |
Resource Type |
How to use |
|---|---|---|---|---|---|
A service for finding mappings (or cross-references) between terms from ontologies, vocabularies and coding standards. |
Ontology alignment |
GUI and API |
Related tools in Bio.Tools
Tool |
Description |
License |
Topics |
Type |
|---|---|---|---|---|
A set of annotation maps describing the entire MeSH assembled using data from MeSH. |
Medical informatics, Data quality management |
Command-line tool, Library |
||
Manually curated database with experimental annotations for the subcellular localizations of proteins in Homo sapiens (HS, human) and Arabidopsis thaliana (AT, thale cress). |
N/A |
Ontology and terminology, Data submission, annotation and curation, Proteins |
Database portal |
7.5.3. Ontology management¶
The process of managing ontologies and other vocabularies in semantic web-linked data environments.This includes policies for update and maintenance of constituent and new terms.
Curated tools
Tool |
Description |
License |
Topics |
Resource Type |
How to use |
|---|---|---|---|---|---|
a repository for biomedical ontologies that aims to provide a single point of access to the latest ontology versions. |
Ontology and terminology |
Web Application, API |
|||
A repository of biomedical ontologies. |
Ontology and terminology |
Web Application, API |
|||
Knowledge Engineering & Knowledge Graph Management. Taxonomy, ontology and linked dataset management. |
Commercial license |
Ontology and terminology |
|||
A centralised, enterprise-ready resource for ontology management and transforms the experience of maintaining and releasing ontologies for research-led businesses. |
Commercial license |
Web application, API |
|||
A linked data server designed for ontologies. Ontobee is aimed to facilitate ontology data sharing, visualization, query, integration, and analysis. |
Web application |
||||
A framework for ontology-based access to biological data. It consists of a repository of bio-ontologies, a set of webservices which provide access to OWL(-EL) reasoning over the ontologies, and several frontends which utilise the ontology repository and reasoning services. |
Web application, API |
Related tools in Bio.Tools
Tool |
Description |
License |
Topics |
Type |
|---|---|---|---|---|
A repository for the NGLY1 Deficiency Knowledge Graph, the reasoning context to support hypothesis discovery for NGLY1 Deficiency-CDDG (DOID:0060728) research. The user can navigate the knowledge in the graph in the Neo4j Browser website. |
N/A |
Molecular interactions, pathways and networks, Ontology and terminology, Machine learning |
Command-line tool |
|
Web application for efficient and accurate Human Phenotype Ontology (HPO) concept curation. |
Genotype and phenotype, Ontology and terminology, Natural language processing |
Web application |
||
A user-friendly interactive tool for the curation and annotation of planarian morphologies and gene expression patterns in a centralized database. |
N/A |
Mathematics, Genotype and phenotype, Model organisms, Ontology and terminology, Gene expression |
||
Advances in gene ontology utilization improve statistical power of annotation enrichment. |
N/A |
Mapping, Ontology and terminology, Microarray experiment |
Command-line tool |
|
This is a project that brings Semantic Web features to the popular Systems Biology software Cytoscape. It allows to query, visualize and reason on ontologies represented in OWL or RDF within Cytoscape. |
Systems biology, Ontology and terminology, Biology |
Desktop application |
||
The tool was developed to manage ontologies (and controlled terminologies e.g. CDISC SEND). The primary goal of the tool is to provide an online collaborative solution for expert curators to map code list terms (sourced from multiple systems/databases) to preferred ontology terms. |
Ontology and terminology, Data identity and mapping |
Web API, Web application |
||
A fast browser for Gene Ontology terms and annotations. |
Ontology and terminology |
Web application |
||
Visualises non-redundant two-level hierarchically structured ontology terms from gene expression data in a 2D space. |
Ontology and terminology, Data visualisation, Gene expression |
Command-line tool, Desktop application |
7.5.4. Ontology engineering¶
Ontology engineering is the process of developing and maintaining ontologies during the ontology life cycle.
Curated tools
Tool |
Description |
License |
Topics |
Resource Type |
|---|---|---|---|---|
A free, open source ontology editor and a knowledge management system. |
Web application, Desktop application |
|||
An open source library and command-line tool for automating ontology development tasks. ROBOT provides ontology processing commands for a variety of tasks, including commands for converting formats, running a reasoner, creating import modules, running reports, and various other tasks. |
Command-line tool |
|||
A Java API and reference implmentation for creating, manipulating and serialising OWL Ontologies. |
LGPL and Apache |
API |
||
A slim tool to slim ontologies as part of ontology integration. It allows users to provide configuration files that specify which parts of an ontology should be kept and/or removed, allowing to just select parts of the ontology you like. |
Java |
|||
TopBraid Composer Maestro Edition is used to develop ontology models, configure data source integration, and create semantic services and user interfaces. |
||||
a web-based, multilingual, collaborative development platform for managing OWL ontologies, SKOS(/XL) thesauri, Ontolex-lemon lexicons and generic RDF datasets. |
Desktop application |
7.6. Implementation examples¶
To show how these tools can be used in real life examples, please check the related recipes.
7.7. References¶
References
7.7.1. What to read next?¶

FAIRsharing records appearing in this recipe:
- BioPortal
- CDISC
- CDISC Standard for Exchange of Nonclinical Data (CDISC SEND)
- Chemical Entities of Biological Interest (ChEBI)
- Comma-separated Values (CSV)
- Disease Ontology (DOID)
- Europe PubMed Central (Europe PMC)
- Gene Ontology (GO)
- Human Phenotype Ontology (HP)
- Monarch Disease Ontology (MONDO)
- Ontobee
- Ontology Cross Reference Service (OxO)
- Ontology Lookup Service (OLS)
- Resource Description Framework (RDF)
- Schema.org
- Simple Knowledge Organization System (SKOS)
- The FAIR Principles (FAIR)
- Web Ontology Language (OWL)
- bio.tools
7.8. Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
EMBL-EBI |
|
Writing - Original Draft, Software |
|||
Barcelona Supercomputing Centre |
Writing - Original Draft, Software |
||||
Sukhi Singh |
The Hyve |
Data curation, Software |
|||
University of Oxford |
Writing - Review & Editing |



