4. Declaring data permitted uses¶

4.1. Main Objectives¶
The purpose of this content is to provide guidance on how to describe permitted use of data and identify the resources that exist to do so.
The aim is also to document equivalent representations and how bridges can be built between the distinct but equivalent implementations.
Finally, the content aims to highlight key use-cases which require coverage, how to code such information, documenting implementation patterns in the context of data cataloguing efforts, for instance by expressing Data Access Policies.
4.3. Tools¶
4.3.1. Standards¶
Data Formats |
Terminologies |
Models |
|---|---|---|
JSON-LD |
||
EGA XML |
||
4.3.2. Implementation¶
4.4. Introduction¶
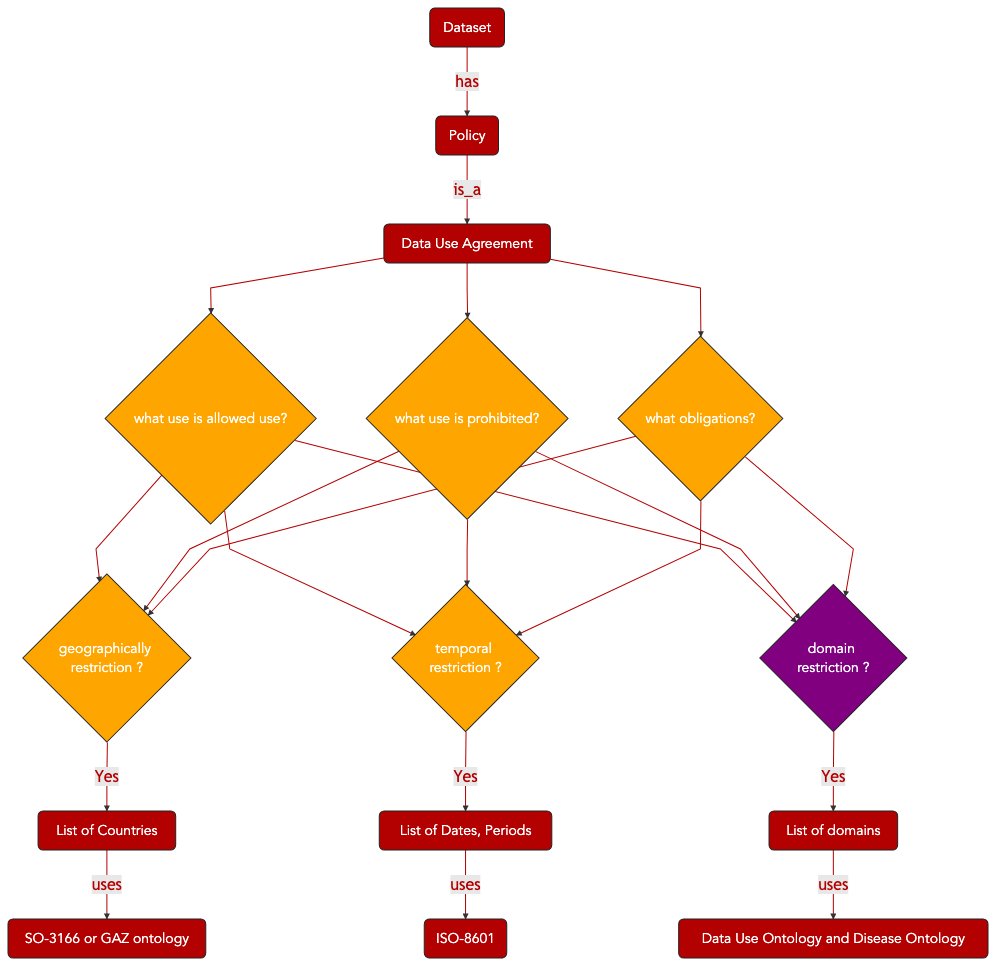
The preservation of patient privacy and the compliance with patient consent are essential considerations when managing sensitive information such as clinical and patient data. Consent forms, as signed by patients, define the acceptable usage of data derived from a patient for research applications. All major research organizations, at national and international levels, enforce strict rules for the management of such data. Sensitive data cannot be accessed without undergoing a vetting process involving a data access request to a data access committee, which will decide, whether or not, to grant requesters access to the data.
This is a time-consuming process in the absence of machine-readable version of data access/ data management policies. In turns, it can prove detrimental to research. Therefore, efforts to enable the provision of concise, efficient and machine processable summary of key permissions and prohibitions have been made. Several resources are now available for the coding and exchange of machine-actionable, legally binding and explicit information related to allowed and consented data usage.
The following sections detail how the international sequence data archives (US NCBI dbGAP, SRA and EU EMBL_EBI EGA) are encoding Data Use Information but also how ODRL, a W3C specification, can be used to represent equivalent information in a format compatible with the data cataloguing efforts relying on W3C DCAT specifications.
4.4.1. SRA & EGA XML schema for Policy, Dataset and Controller¶
Next Generation Sequencing (NGS) techniques allow routine production of full genome data from patients. This data is highly sensitive and data repositories specialized in storing such information have developed procedures and representation models for defining the conditions of use.
We summarize here the key objects used by the European Genome Archive, in compliance with INSDC and GA4GH guidelines.
https://ega-archive.org/data-use-conditions
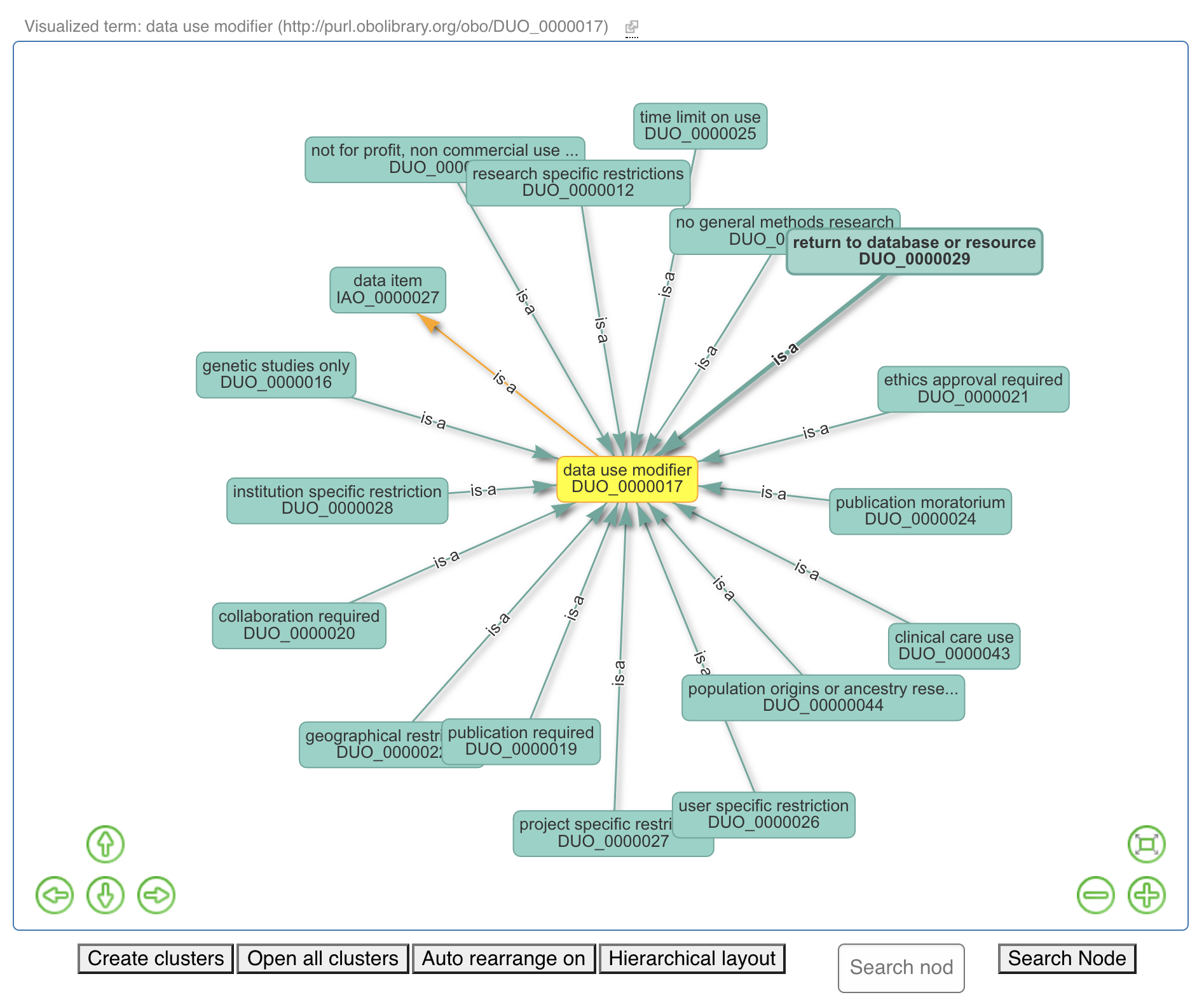
Fig. 4.7 Data Use Ontology Overview Part 1¶

Fig. 4.8 Data Use Ontology Overview Part 2¶
The information presented below has been sourced from the ENA GitHub repo.
The Data Access Committee and Contact information
<?xml version = '1.0' encoding = 'UTF-8'?>
<DAC_SET>
<DAC alias="DAC-2011-08-11T11:45:28Z-1873" accession="EGAC00000000001" center_name="EBI" broker_name="EGA">
<IDENTIFIERS>
<PRIMARY_ID>EGAC00000000001</PRIMARY_ID>
<SUBMITTER_ID namespace="EBI">DAC-2011-08-11T11:45:28Z-1873</SUBMITTER_ID>
</IDENTIFIERS>
<TITLE>EGA DAC TITLE</TITLE>
<CONTACTS>
<CONTACT name="Joe Bloggs" email="joe@noname.com" organisation="EBI"/>
</CONTACTS>
</DAC>
</DAC_SET>
The Data Access Policy object
Note
in the following example, the text of the policy is present in the XML representation
<?xml version = '1.0' encoding = 'UTF-8'?>
<POLICY_SET>
<POLICY center_name="EBI" alias="Policy-2011-08-26T12:23:53Z-1868" accession="EGAP00001000001" broker_name="EBI">
<IDENTIFIERS>
<PRIMARY_ID>EGAP00001000001</PRIMARY_ID>
<SUBMITTER_ID namespace="SC">Policy-2011-08-26T12:23:53Z-1868</SUBMITTER_ID>
</IDENTIFIERS>
<TITLE/>
<DAC_REF accession="EGAP00001000001" refname="DAC_-2011-08-26T12:23:49Z-1868" refcenter="EBI">
<IDENTIFIERS>
<PRIMARY_ID>EGAC00001000001</PRIMARY_ID>
<SUBMITTER_ID namespace="EBI">DAC_-2011-08-26T12:23:49Z-1868</SUBMITTER_ID>
</IDENTIFIERS>
</DAC_REF>
<POLICY_TEXT>https://www.sanger.ac.uk/datasharing/</POLICY_TEXT>
</POLICY>
</POLICY_SET>
Note
In the following example, the file address (url) to the policy is included in the XML representation. Ideally, the url provided should be a globally unique persistent identifier so one can be sure to obtain at least the metadata about the document.
<?xml version = '1.0' encoding = 'UTF-8'?>
<POLICY_SET>
<POLICY center_name="EBI" alias="Policy-2011-08-26T12:23:53Z-1868" accession="EGAP00001000001" broker_name="EBI">
<IDENTIFIERS>
<PRIMARY_ID>EGAP00001000001</PRIMARY_ID>
<SUBMITTER_ID namespace="SC">Policy-2011-08-26T12:23:53Z-1868</SUBMITTER_ID>
</IDENTIFIERS>
<TITLE/>
<DAC_REF accession="EGAP00001000001" refname="DAC_-2011-08-26T12:23:49Z-1868" refcenter="EBI">
<IDENTIFIERS>
<PRIMARY_ID>EGAC00001000001</PRIMARY_ID>
<SUBMITTER_ID namespace="EBI">DAC_-2011-08-26T12:23:49Z-1868</SUBMITTER_ID>
</IDENTIFIERS>
</DAC_REF>
<POLICY_FILE>https://www.sanger.ac.uk/datasharing/</POLICY_FILE>
</POLICY>
</POLICY_SET>
Expressing Data Use with EGA XML and Data Use Ontology codes.
<?xml version = '1.0' encoding = 'UTF-8'?>
<POLICY_SET>
<POLICY center_name="EBI" alias="Policy-2011-08-26T12:23:53Z-1868" accession="EGAP00001000001" broker_name="EBI">
<IDENTIFIERS>
<PRIMARY_ID>EGAP00001000001</PRIMARY_ID>
<SUBMITTER_ID namespace="SC">Policy-2011-08-26T12:23:53Z-1868</SUBMITTER_ID>
</IDENTIFIERS>
<TITLE/>
<DAC_REF accession="EGAP00001000001" refname="DAC_-2011-08-26T12:23:49Z-1868" refcenter="EBI">
<IDENTIFIERS>
<PRIMARY_ID>EGAC00001000001</PRIMARY_ID>
<SUBMITTER_ID namespace="EBI">DAC_-2011-08-26T12:23:49Z-1868</SUBMITTER_ID>
</IDENTIFIERS>
</DAC_REF>
<POLICY_FILE>https://www.sanger.ac.uk/datasharing/</POLICY_FILE>
<DATA_USES>
<!-- no restriction -->
<DATA_USE>http://purl.obolibrary.org/obo/DUO_0000004</DATA_USE>
<DATA_USES>
<DATA_USE ontology="DUO" code="0000004" version="17-07-2016"/>
</DATA_USES>
</DATA_USES>
</POLICY>
</POLICY_SET>
Indicating disease specific restriction on research with DUO and ontologies covering the Disease and Pathology domain.
<?xml version = '1.0' encoding = 'UTF-8'?>
<POLICY_SET>
<POLICY center_name="EBI" alias="Policy-2011-08-26T12:23:53Z-1868" accession="EGAP00001000001" broker_name="EBI">
<IDENTIFIERS>
<PRIMARY_ID>EGAP00001000001</PRIMARY_ID>
<SUBMITTER_ID namespace="SC">Policy-2011-08-26T12:23:53Z-1868</SUBMITTER_ID>
</IDENTIFIERS>
<TITLE/>
<DAC_REF accession="EGAP00001000001" refname="DAC_-2011-08-26T12:23:49Z-1868" refcenter="EBI">
<IDENTIFIERS>
<PRIMARY_ID>EGAC00001000001</PRIMARY_ID>
<SUBMITTER_ID namespace="EBI">DAC_-2011-08-26T12:23:49Z-1868</SUBMITTER_ID>
</IDENTIFIERS>
</DAC_REF>
<POLICY_FILE>https://www.sanger.ac.uk/datasharing/</POLICY_FILE>
<DATA_USES>
<!-- ethics approval required -->
<DATA_USE>http://purl.obolibrary.org/obo/DUO_0000021</DATA_USE>
<!-- geographical restriction -->
<DATA_USE>http://purl.obolibrary.org/obo/DUO_0000022</DATA_USE>
<!-- not-for-profit-organization-use-only -->
<DATA_USE>http://purl.obolibrary.org/obo/DUO_0000045</DATA_USE>
<!-- disease specific research -->
<DATA_USE>http://purl.obolibrary.org/obo/DUO_0000007</DATA_USE>
</DATA_USES>
</POLICY>
</POLICY_SET>
Note
When using the consent code DUO_0000007 where data is restricted for use on a specific disease area, it is necessary to explicitly indicate which disease area is allowed. This can be done by associating codes/identifiers from well established disease terminologies such as MONDO, DOID, SNOMED-CT.
For instance, if data reuse is restricted to research into juvenile idiopathic arthritis, the code should be displayed as DUO_0000007; MONDO:0011429
<POLICY_SET>
<POLICY alias="ena-POLICY-BABRAHAM-23-03-2017-09:47:38:853-62" center_name="BABRAHAM" accession="EGAP00001000615" broker_name="EGA">
<IDENTIFIERS>
<PRIMARY_ID>EGAP00001000615</PRIMARY_ID>
<SUBMITTER_ID namespace="BABRAHAM">ena-POLICY-BABRAHAM-23-03-2017-09:47:38:853-62</SUBMITTER_ID>
</IDENTIFIERS>
<TITLE>Data Access Agreement for PCHiC, RNA-Seq, ChIP-Seq</TITLE>
<DAC_REF accession="EGAC00001000523">
<IDENTIFIERS>
<PRIMARY_ID>EGAC00001000523</PRIMARY_ID>
</IDENTIFIERS>
</DAC_REF>
<POLICY_FILE>ftp://ftp.ebi.ac.uk/pub/contrib/pchic/EGA_Data_Access_Request_DIL.docx</POLICY_FILE>
<DATA_USES>
<DATA_USE ontology="DUO" code="0000007" version="17-07-2016">
<!-- disease specific research -->
<MODIFIER>
<DB>EFO</DB>
<ID>0001645</ID>
</MODIFIER>
<MODIFIER>
<DB>EFO</DB>
<ID>0001655</ID>
</MODIFIER>
</DATA_USE>
<DATA_USE ontology="DUO" code="0000014" version="17-07-2016"/>
</DATA_USES>
</POLICY>
</POLICY_SET>
<DATASETS>
<DATASET alias="EGAS000000001-sc-20110919" center_name="SC" broker_name="EGA" accession="EGAD00001000039">
<IDENTIFIERS>
<PRIMARY_ID>EGAD00001000039</PRIMARY_ID>
<SUBMITTER_ID namespace="SC">EGAS000000001-sc-20110919</SUBMITTER_ID>
</IDENTIFIERS>
<TITLE>Platelet collagen defect</TITLE>
<RUN_REF accession="EGAR0000000001" refname="RUN_1" refcenter="EBI">
<IDENTIFIERS>
<PRIMARY_ID>EGAR0000000001</PRIMARY_ID>
<SUBMITTER_ID namespace="EBI">RUN_1</SUBMITTER_ID>
</IDENTIFIERS>
</RUN_REF>
<POLICY_REF accession="EGAP00000001" refname="Policy_-2011-08-17T15:05:39Z-1888" refcenter="EBI">
<IDENTIFIERS>
<PRIMARY_ID>EGAP00001000024</PRIMARY_ID>
<SUBMITTER_ID namespace="EBI">Policy_-2011-08-17T15:05:39Z-1888</SUBMITTER_ID>
</IDENTIFIERS>
</POLICY_REF>
</DATASET>
</DATASETS>
Browsing Data Access Committees available from EGA:
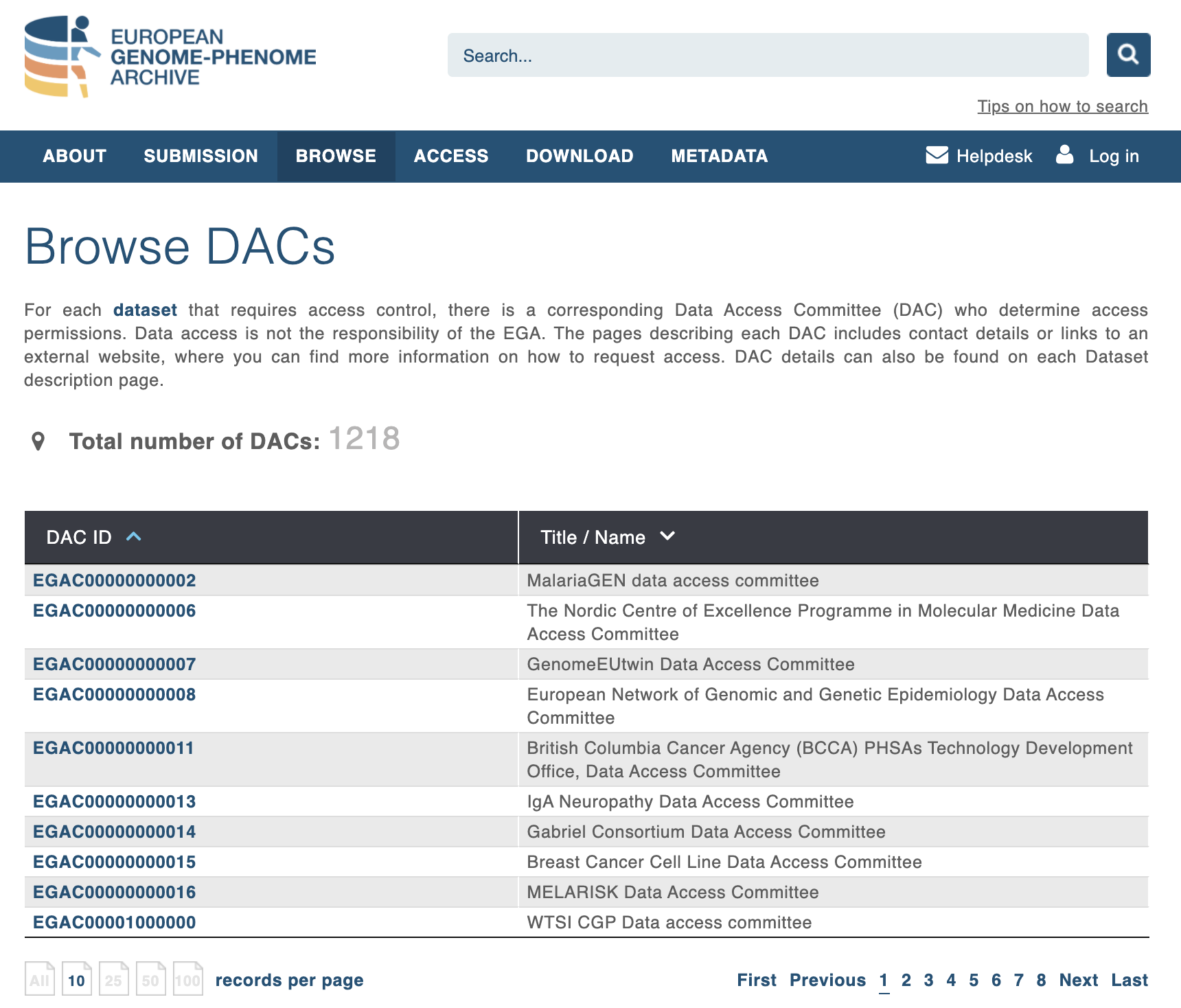
Fig. 4.9 List of Data Access Policy from EGA¶
4.4.2. ODRL, Open Digital Rights Language¶
ODRL stands for Open Digital Rights Language and is a set of W3C Recommendations defining a policy expression language.
ODRL is made up of several components:
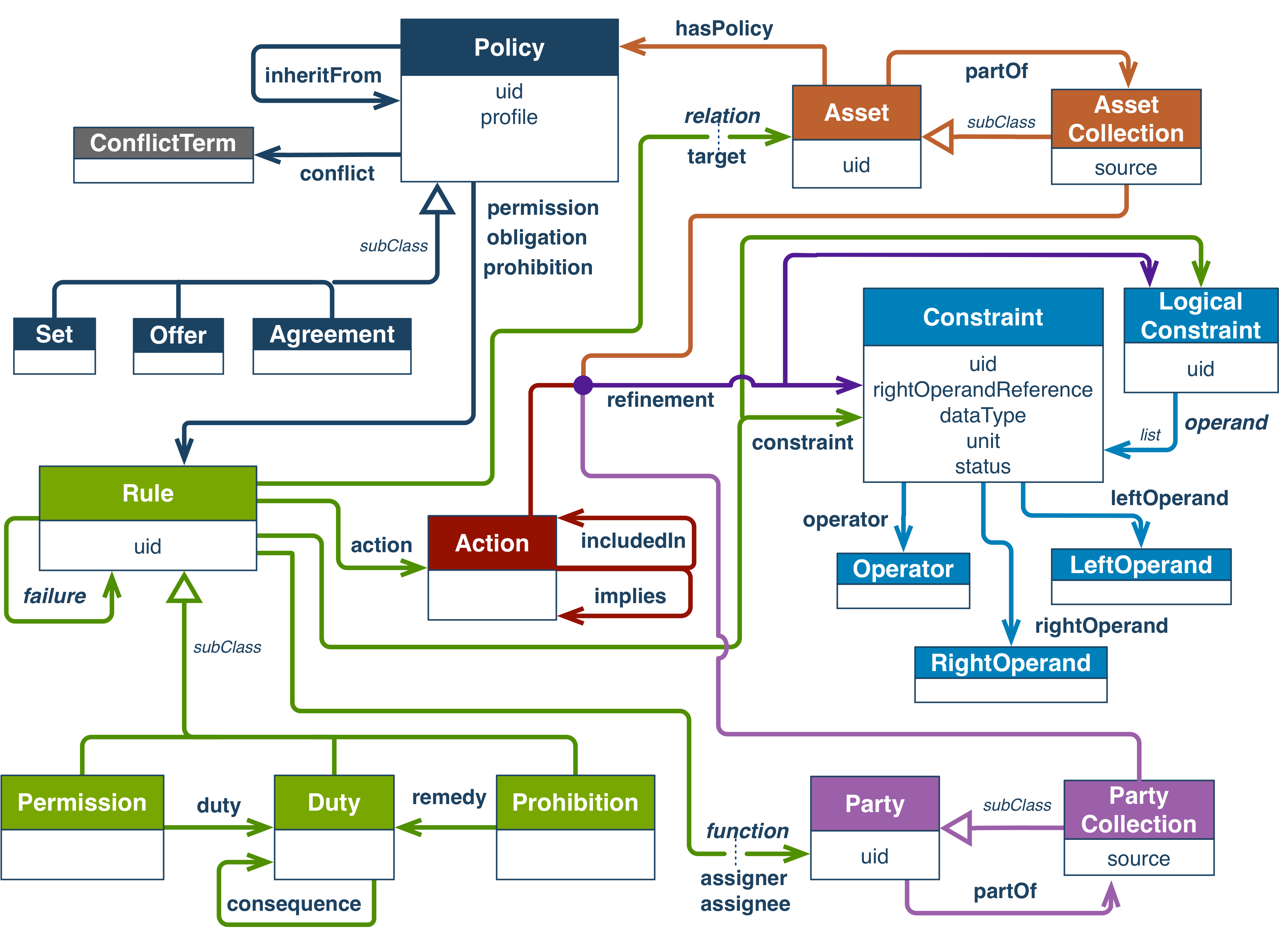
Fig. 4.10 ODRL overview¶
The ODRL Vocabulary and Expression provides the terms to express policies in RDF language.
The ODRL Vocabulary and Expression complements the ODRL information model, which allows expressing similar information in JSON language.
Warning
In 2015, the dedicated working group produced the following JSON schema implementation guidance https://www.w3.org/community/odrl/json/2.1/#section-Schema
We base our representations on this specification 2.
We are aware of a possible misalignment between the specifications of the Working Group (from 2015) and the latest specifications as to whether to use the keys “name” or “leftOperand” (https://www.w3.org/TR/odrl-model/#constraint-rule, 2018). In the following representations, we use the key “name” to validate against the 2015 JSON-schema https://www.w3.org/community/odrl/json/2.1/#section-Schema / https://github.com/iptc/rightsml-dev/blob/master/licensed/ODRL21.json
4.4.2.1. The different types of Policies¶
The ODRL model defines several subclasses for the Policy entity, namely Agreement, Set, Offer.
4.4.2.1.1. Describing an agreement with ODRL¶
{
"policytype": "http://www.w3.org/ns/odrl/2/Agreement",
"policyid": "http://example.com/policy:5531",
"inheritallowed": true,
"permissions": [{
"target": "http://example.com/report:2321",
"action": "http://www.w3.org/ns/odrl/2/print",
"assigner": "http://example.com/pub:88",
"assignee": "http://example.com/billie:888"
}]
}
{
"policytype": "http://www.w3.org/ns/odrl/2/Agreement",
"policyid": "http://example.com/policy:9999",
"inheritfrom": "http://example.com/policy:5531",
"permissions": [{
"target": "http://example.com/report:2333",
"action": "http://www.w3.org/ns/odrl/2/display",
"assigner": "http://example.com/pub:88",
"assignee": "http://example.com/class:IT01",
"assignee_scope": "http://www.w3.org/ns/odrl/2/group"
}]
}
4.4.3. Encoding Research Restriction on disease and geographical area using ODRL and DUO¶
In this section, we document how to rely on a basic ODRL-based pattern using a DUO term 1 to represent a situation where SecondaryUse of the data is allowed on the condition that work is restricted to disease specific research.
{
"policytype": "http://www.w3.org/ns/odrl/2/Policy",
"policyid": "https://fairplus.github.io/examples/policy_122334",
"permissions": [
{
"target": "https://fairplus.github.io/examples/dataset_00001",
"action": "http://www.w3.org/ns/odrl/2/secondaryUse",
"assigner": "https://fairplus.github.io/examples/examples/efpia_organization_00002",
"constraints":[{
"name": "http://www.w3.org/ns/odrl/2/purpose",
"operator": "http://www.w3.org/ns/odrl/2/eq",
"rightoperand": "http://purl.obolibrary.org/obo/DUO_0000007"
}
]
}
]
}
Note
The main limitation of the representation is that it provides no information about which diseases are vetted for research.
The following representation is more sophisticated and includes 3 types of restrictions:
restriction on specific disease (juvenile arthritis, to reuse the exemplar representation in EGA/SRA XML presented in section 1)
restriction on the geographical location where the research can be conducted
an obligation to delete the data obtained through the access agreement past a specified duration, 3 years in our example
Let’s proceed stepwise.
Representing Research Restriction on Specific Disease Area using DUO and MONDO ontologies
{
"policytype": "http://www.w3.org/ns/odrl/2/Policy",
"policyid": "https://fairplus.github.io/examples/policy_122334",
"permissions": [
{
"target": "https://fairplus.github.io/examples/dataset_00001",
"action": [{
"rdf:value": { "@id": "odrl:secondaryUse" },
"refinement": {
"xone": {
"@list": [
{ "@id": "http://purl.obolibrary.org/obo/MONDO_0011429" },
{ "@id": "http://purl.obolibrary.org/obo/EFO_0001645" },
{ "@id": "http://purl.obolibrary.org/obo/EFO_0001655" }
]
}
}
}],
"assigner": "https://fairplus.github.io/examples/examples/efpia_organization_00002",
"constraints":[{
"name": "http://www.w3.org/ns/odrl/2/purpose",
"operator": "http://www.w3.org/ns/odrl/2/eq",
"rightoperand": "http://purl.obolibrary.org/obo/DUO_0000007"
}
]
}
]
}
Note
When using refinements, note the difference in representation to indicate the nature of the action. here it uses:
while the ordinary is simpler: “action”: “http://www.w3.org/ns/odrl/2/secondaryUse”,
vs
“action”: [{ “rdf:value”: { “@id”: “odrl:secondaryUse” },
Note
While DUO is unique in its coverage of data uses, various disease ontologies exist and may be used to specify the specific focus research should have. For instance, SNOMED-CT, Disease Ontology could also be used. It is worth noting that extensive cross referencing exists between resources such as DOID, MONDO and SNOMED-CT but this is something to consider when implementing brokering systems.
Representing Research Restriction based on Geographical Regions
The section shows how to use ODRL to document geographical restrictions, either by listing countries where research is allowed or by listing those countries excluded from doing so.
In the following example, research is only allowed in a specific country, Italy in this case, which is encoded using the ISO-3166 code.
{
"policytype": "http://www.w3.org/ns/odrl/2/Policy",
"policyid": "https://fairplus.github.io/examples/policy_122334",
"permissions": [
{
"target": "https://fairplus.github.io/examples/dataset_00001",
"action": [{
"rdf:value": { "@id": "odrl:secondaryUse" },
"refinement": {
"xone": {
"@list": [
{ "@id": "http://purl.obolibrary.org/obo/MONDO_0011429" },
{ "@id": "http://purl.obolibrary.org/obo/EFO_0001645" },
{ "@id": "http://purl.obolibrary.org/obo/EFO_0001655" }
]
}
}
}],
"assigner": "https://fairplus.github.io/examples/examples/efpia_organization_00002",
"constraints":[{
"name": "http://www.w3.org/ns/odrl/2/purpose",
"operator": "http://www.w3.org/ns/odrl/2/eq",
"rightoperand": "http://purl.obolibrary.org/obo/DUO_0000007"
},
{
"name": "http://www.w3.org/ns/odrl/2/spatial",
"operator": "http://www.w3.org/ns/odrl/2/eq",
"rightoperand": "http://www.itu.int/tML/tML-ISO-3166:it"
}
]
}
]
}
Representing Obligations regarding Data Management
The following example shows how to explicitly state in a Policy element that the data must be deleted after a defined period of time (3 years in this example). Duration and time related value should be represented using ISO-8601 standard.
{
"policytype": "http://www.w3.org/ns/odrl/2/Policy",
"policyid": "https://fairplus.github.io/examples/policy_122334",
"permissions": [
{
"target": "https://fairplus.github.io/examples/dataset_00001",
"action": "http://www.w3.org/ns/odrl/2/secondaryUse",
"action": [{
"rdf:value": { "@id": "odrl:secondaryUse" },
"refinement": {
"xone": {
"@list": [
{ "@id": "http://purl.obolibrary.org/obo/MONDO_0011429" },
{ "@id": "http://purl.obolibrary.org/obo/EFO_0001645" },
{ "@id": "http://purl.obolibrary.org/obo/EFO_0001655" }
]
}
}
}],
"assigner": "https://fairplus.github.io/examples/examples/efpia_organization_00002",
"constraints":[{
"name": "http://www.w3.org/ns/odrl/2/purpose",
"operator": "http://www.w3.org/ns/odrl/2/eq",
"rightoperand": "http://purl.obolibrary.org/obo/DUO_0000007"
},
{
"name": "http://www.w3.org/ns/odrl/2/spatial",
"operator": "http://www.w3.org/ns/odrl/2/eq",
"rightoperand": "http://www.itu.int/tML/tML-ISO-3166:it"
}
],
"duties": [{
"action": "http://www.w3.org/ns/odrl/2/delete",
"target": "https://fairplus.github.io/examples/dataset_00001",
"constraints": [{
"name": "http://www.w3.org/ns/odrl/2/dateTime",
"operator": "http://www.w3.org/ns/odrl/2/eq",
"rightoperand": "P36M"
}]
}]
}
]
}
4.6. Implementation in Data Catalogues built with DCAT or DATS¶
4.6.1. Referring to an ODRL Policy from a DCAT DataSet¶
@prefix dcat:<https://www.w3.org/ns/dcat> .
@prefix odrl:<https://www.w3.org/ns/odrl/2/core> .
@prefix dct: <http://purl.org/dc/elements/1.1/> .
@prefix xsd: <http://www.w3.org/2001/XMLSchema#> .
<#daa-policy-1>
a odrl:Policy .
<#dataset-001>
a dcat:Dataset ;
dct:title "Human Patient Genomic Dataset"@en ;
dcat:keyword "genotype"@en, "phenotyping"@en, "IMI"@en ;
dct:creator "imi-consortium-XYZ" ;
dct:issued "2021-12-05"^^xsd:date ;
dct:modified "2021-12-15"^^xsd:date ;
dcat:contactPoint <http://example.org/imi-project-xyz/contact1> ;
dct:publisher "imi-consortium-XYZ";
dct:language <http://id.loc.gov/vocabulary/iso639-1/en> ;
dcat:distribution _:dataset-001-csv ;
odrl:hasPolicy <#daa-policy-1>;
.
For more details, please see the following publication by de Vos et al, 2019
4.6.2. DATS and ODRL JSON¶
An evolution of the DATS 7, 5 schema is used by University of Luxembourg to build a data catalogue for IMI projects and datasets. The proposed patterns could be used and tested to representation DAC/DAA information as well as the allowed uses of datasets generated by the consortia funded by IMI. More complex use cases can be considered to assess to ability of the representations to be associated to specific datasets, for instance datasets associated with a particular data acquisition technique the access of which may require specific policies and conditions to be made machine-readable.
The approach is therefore to reference the JSON representations compliant with the ODRL JSON schema 2.1 specification, as presented in earlier sections in conjunction with DATS JSON documents.
4.6.3. Validating ODRL RDF documents¶
Code exists that allows developers to validate ODRL documents expressed in legitimate RDF serializations.
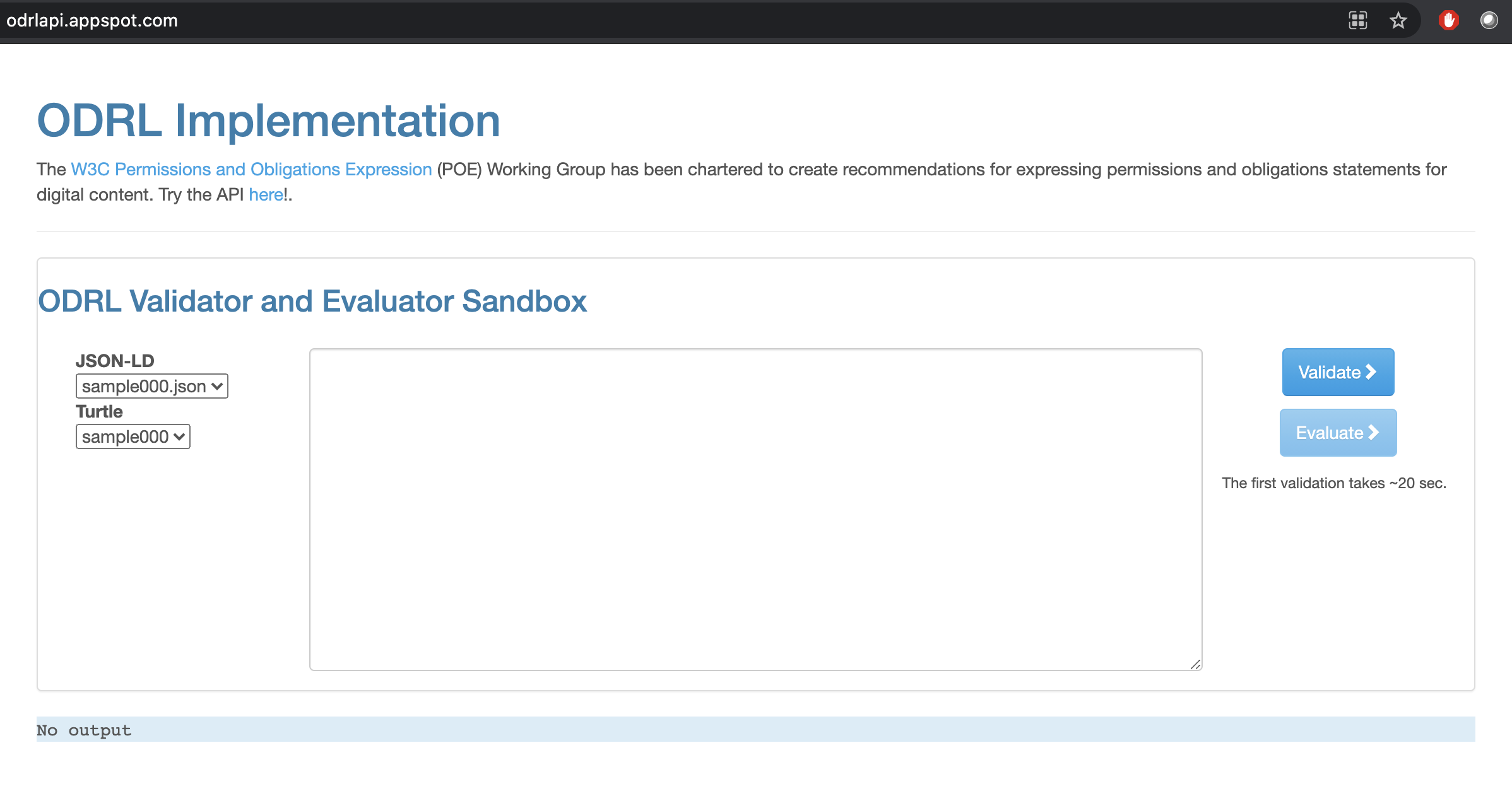
Fig. 4.11 ODRL Validation and Evaluation Sandbox¶
The web-application is powered by a REST-API the swagger document of which is available from the following address: http://odrlapi.appspot.com/apidoc/index.html
4.8. Conclusion¶
Making sure that machine-readable information about the conditions of use of datasets and data is available is key to enable privacy preserving and policy compliant use of information across organizations. This content provides an overview of the models available to do so and how it has been applied to life science data, showing the main features of the models and how to define use based on the major properties such as the type of research allowed, how to indicate disease domain or geographical restrictions as well as temporal restrictions associated to the dataset and defined by the data owners/data controllers.
4.8.1. What to read next?¶

Learn more about:

FAIRsharing records appearing in this recipe:
4.9. References¶
References
- 1
Duo. URL: http://purl.obolibrary.org/obo/duo.owl.
- 2
Odrl json schema. URL: https://www.w3.org/community/odrl/json/2.1/#section-Schema.
- 3(1,2)
Odrl model. URL: https://www.w3.org/TR/odrl-model.
- 4
Odrl vocabulary. URL: https://www.w3.org/TR/odrl-vocab/.
- 5
G. Alter, A. Gonzalez-Beltran, L. Ohno-Machado, and P. Rocca-Serra. The Data Tags Suite (DATS) model for discovering data access and use requirements. Gigascience, 02 2020.
- 6
S. O. Dyke, A. A. Philippakis, J. Rambla De Argila, D. N. Paltoo, E. S. Luetkemeier, B. M. Knoppers, A. J. Brookes, J. D. Spalding, M. Thompson, M. Roos, K. M. Boycott, M. Brudno, M. Hurles, H. L. Rehm, A. Matern, M. Fiume, and S. T. Sherry. Consent Codes: Upholding Standard Data Use Conditions. PLoS Genet, 12(1):e1005772, Jan 2016.
- 7
S. A. Sansone, A. Gonzalez-Beltran, P. Rocca-Serra, G. Alter, J. S. Grethe, H. Xu, I. M. Fore, J. Lyle, A. E. Gururaj, X. Chen, H. E. Kim, N. Zong, Y. Li, R. Liu, I. B. Ozyurt, and L. Ohno-Machado. DATS, the data tag suite to enable discoverability of datasets. Sci Data, 4:170059, 06 2017.
4.10. Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
Tom Plasterer |
Astra-Zeneca |
Conceptualization, Writing, Review & Editing |
|||
University of Oxford |
Conceptualization, Writing - Original Draft, Review & Editing |
||||
EMBL-EBI |
Review & Editing |
||||
EMBL-EBI |
|
Review & Editing |
|||
University of Luxembourg |
Review & Editing |