FAIR and Knowledge graphs¶

Main Objectives¶
In the following sections, we will cover the following topics:
the relation between FAIR and KGs.
the typology of knowledge graphs (KG).
the different technologies supporting them.
the methods to generate knowledge graphs.
the impact of graph-based representations on what it means to deliver FAIR data and services.
FAIR and Knowledge Graphs¶
FAIR is chiefly about three things: metadata, metadata and metadata.
Not only that, but metadata which should be active, that is usable by software agents without the need of human intervention, thanks to the resolvable links found in the electronic documents and the associated semantics available to the agent. The goal is to speed up data handling by ensuring enough information is provided to machines.
In this context, the availability of semantic web technologies completely aligns with the key requirements defined by the FAIR principles. Thus, providing metadata about a dataset in the form of Linked Data Graph is a significant path towards making data FAIR. Furthermore, the availability of knowledge representation in a graph data structure, or knowledge graphs, provides contextual information about the dataset and shows the relationship between data and curated domain knowledge, which is particularly useful for machine learning and artificial intelligence approach.
But
FAIR dataandknowledge graphsare not equivalent. Not all FAIR data is a knowledge graph and not all knowledge graphs are FAIR.
What are knowledge graph and graph databases?¶
Knowledge Graph (KG) and graph databases constitute a new approach to representation, storage and querying of data.
To understand the notion of knowledge graphs, we need to remind ourselves about some elements of information theory, data structure, and data storage, as well as some geometric interpretation of relationship between entities, which is often what turns data into knowledge.
Information, in digital forms, relies on formal representations and an array of methods to store and retrieve it. Up until the last 10 years, Relational DataBase Management Systems (RDBMS) have been provided the backbone of information storage solutions, and still do so today. However, advances in both theoretical information representation models and technical solutions led to the development of so-called NoSQL solutions (for ‘Not Only Structured Query language’), supporting the onset of graph-based representations of information, in contrast to relational-model based representation. Knowledge graphs are knowledge bases which use graph-based data structure or topology to represent entities and the relationships between them.
So how does graph theory come to meet knowledge representation?
We need here to introduce the definition of a graph:
A graph is a set of nodes (also known as vertex) connected via edges.
When the edges connected any two nodes carry a direction information, they defined a specific and important subfamily of graphs known as directed graphs. On the other hand, if there is absence of directionality information on the edges, such graphs are known as undirected graphs.
A graph can also contain cycles. This means that when traversing the graph from a starting or source node to an ending or target node (terminologically referred to as a path), in case of cycles, the source and the target node are the same. Such graphs are commonly known as cyclic graphs.
When cycles aren’t allowed, a new subtype of graph is defined. Such graphs are known as directed acyclic graphs (DAG).
Volumes have been written about what graphs are and the underlying mathematical theories describing their properties. The readership can refer to the following resources for a complete overview and training. The purpose of this section is simply to provide a rapid overview of what knowledge graphs are and their typology, as one needs to be aware of the differences that exist between the different kinds of graphs and also understand the consequences both theoretical and technical to fully appreciate the pros and cons of the various implementations available out there.
The following sections intend to provide a compact review and guide newcomers through the jargon and the various acronyms founds in the literature on the topic.
Relational Databases and Graph Databases¶
Until fairly recently, the storage of information mainly relied on RDBMS,
which have been to work-horse of database building. With relational database approach, information is modelled according to an Entity Relationship Model (ER model/ER diagram), the development of which undergoes a series of formal steps.
One of these steps involves a process known as schema normalization to ensure that key entities and their attributes can be stored in tables in the most parsimonious way. This step is necessary to ensure query performance and optimization.
Relational database approaches track and store relations between entities using what is known as linking tables. When interrogating a database operating under this paradigm (e.g. MySQL, PostgreSQL, Oracle DB, to name a few), the database engine needs to run JOIN operations, a type of SQL queries hitting the table of entities and any linking table holding the information about the relations between entities.
While sophisticated optimization methods exist to ensure query performance does not suffer as both data volume grows or complexity of the underlying model increases, this form of information storage may suffer from performance issue in certain situations.
An added criticism often leveled to RDBMS based database is the rigidity, which manifests itself in the difficulty and complexity of changing the underlying model, leading to potentially complex migration tasks. This can become a limitation when knowledge representation in a particular domain requires frequent changes for whatever reason.
So it is in this context that the advent of graph (oriented) databases (sometimes referred to as NoSQL database, for ‘Not Only SQL database’), came about, with a new paradigm and bold claims.
Over the last 20 years, the idea of storing knowledge as a graph G=(v for vertex, e for edge),
even if not new at all from a theoretical perspective, has undergone tremendous progress to the point that a transition,
in some sectors of the industry, is taking place.
KGs and graph databases claim to be capable of offering new insights with better performance, owing to the optimization of their query engine for fast traversals. With description logics (DL), KGs represent knowledge to allow “intelligent agents”, machines and algorithms, to reason over and work with. Hence, for some domain specific representations and tasks, KG and graph databases are seen as more suited than RDBMS storage.
These benefits warrant the significant investments made to develop tools and frameworks to support KG storage solutions. It also means that KG are becoming mainstream.
It is not that graph-oriented data storage is replacing RDBMS storage. As always, with all technologies and tooling, one needs to understand the task at hand and choose the most suitable solutions for addressing a specific challenge.
The different types of knowledge graphs¶
The 2 main types of Knowledge Graph - Graphical Overview
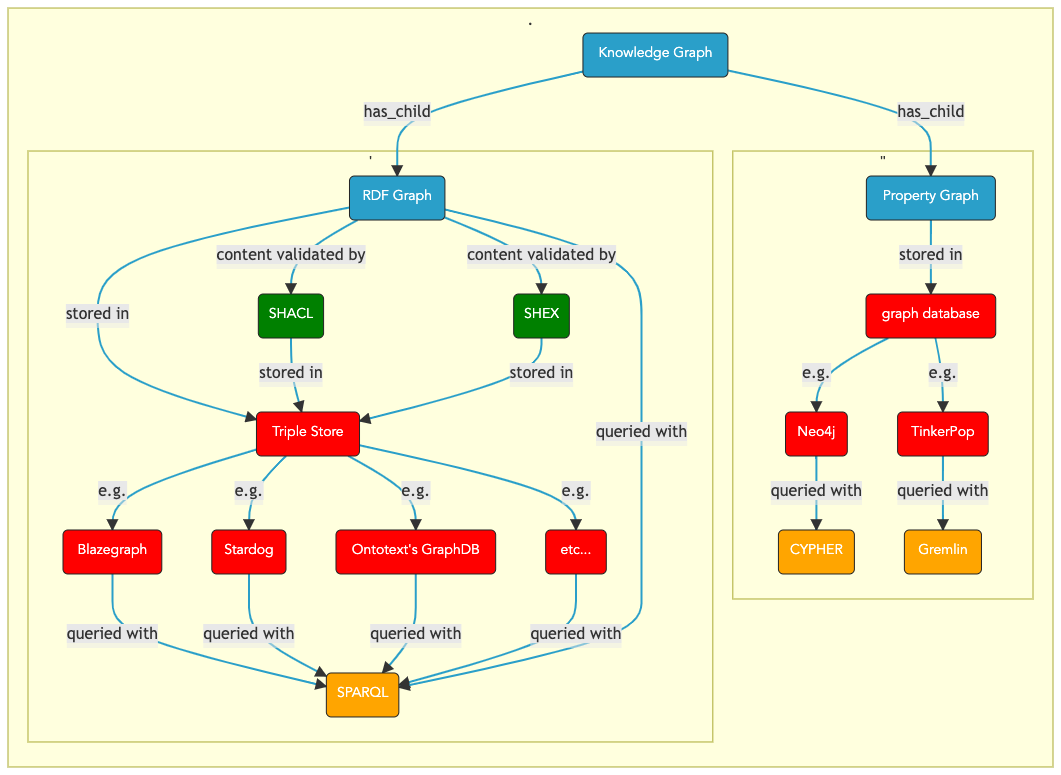
Fig. 8 Data Use¶
RDF Graph and Labeled Property graphs
1. RDF graphs¶
i. Defining RDF, Resource Description Framework¶
Here, we need to introduce the notion of representing statements as a predicate of the following form:
<subject><predicate><object>, such as: 'P53 protein' 'interacts_with' 'DNA'.
This statement can be formally expressed using a dedicated syntax called RDF,
standing for “Resource Description Framework”, one of the World Wide Web Consortium (W3C) standards supporting the vision of a Semantic Web as
outlined by Sir Tim Berners-Lee and colleagues 3.
In this instance, because the relation 'interacts_with' is directional, we are creating the simplest form of directed graph,
by establishing an edge between two nodes.
When does a RDF Graph become a Linked Data Graph?
When all the entities in an RDF statement are identified and available as Universal Resource Identifiers(URI),
the RDF graph then becomes a Linked Data Graph, since each entity is no longer an plain literal and can now be
looked up by means of HTTP requests .
`P53 protein` `physically interacts with` `DNA`.
using Wikidata identifiers can be expressed as:
"@wdt": "https://www.wikidata.org/wiki/"
[wdt:Q283350](https://www.wikidata.org/wiki/Q283350) [wdt:Property:P129](https://www.wikidata.org/wiki/Property:P129)
[wdt:Q7430](https://www.wikidata.org/wiki/Q7430)
The RDF syntax allows the expression of RDF statements, which can be built and grouped in an RDF graph.
So let’s go back to the statement about the gene product of the P53 gene. The formal and structured representation of a fact about that gene is now a well established technique. Therefore, one can see that natural fit between the data structure and knowledge gathered and accumulated by scientists. This is why knowledge graphs have gained so much traction in recent years. In fact, a number of developments converged to make this form of representation effective and above all useful for computational work.
ii. Persisting RDF graphs: RDF triple Stores¶
RDF graph objects can be persisted in specialized databases, RDF graph databases also known as RDF triple stores.
Some of the most performant and successful solutions are:
iii. Validating and controlling the quality of the RDF data being loaded -¶
To validate and control the quality of the fast emerging RDF triples.
The W3C has produced a specification detailing a constraint language which allows data managers to control the so-called
shape of the RDF graph coming in. The W3C SHACL SHApe Constraint Language, known as SHACL (pronounced shackle),
allow to express a set of conditions to validate RDF graphs/RDF statements. SHACL expressions are RDF statements
and the constraint profiles can be stored in an RDF triple store.
SHACL specifications are implemented in the TopQuadrant TopBraid Composer tool.
A competing specification, know as SHEX for Shape Expression, provides a similar functionality,
but isn’t a W3C approved specifications, even though SHEX is proving quite popular and with a strong following.
iv. Querying an RDF graph - the W3C SPARQL Query Language¶
RDF graphs stored in RDF triple stores can be queried using a dedicated query language defined by a W3C specification known as
SPARQL 1.1.
SPARQL stands for SPARQL Query Language, (pronounced sparkle) 1.
The results of a SPARQL query are a result set or RDF graph and is therefore a collection of RDF triples.
An impressive feature of the SPARQL query language is its ability to perform mashups by performing federated queries
over a number of ‘SERVICES’, i.e. RDF triple store endpoints and return an RDF graph which contains triples assembled
from a number of resources.
We still need to introduce several key concepts to provide a fuller picture of knowledge graph, how they are generated and why they matter.
SPARQL query example: Metabolites and the species they are found in
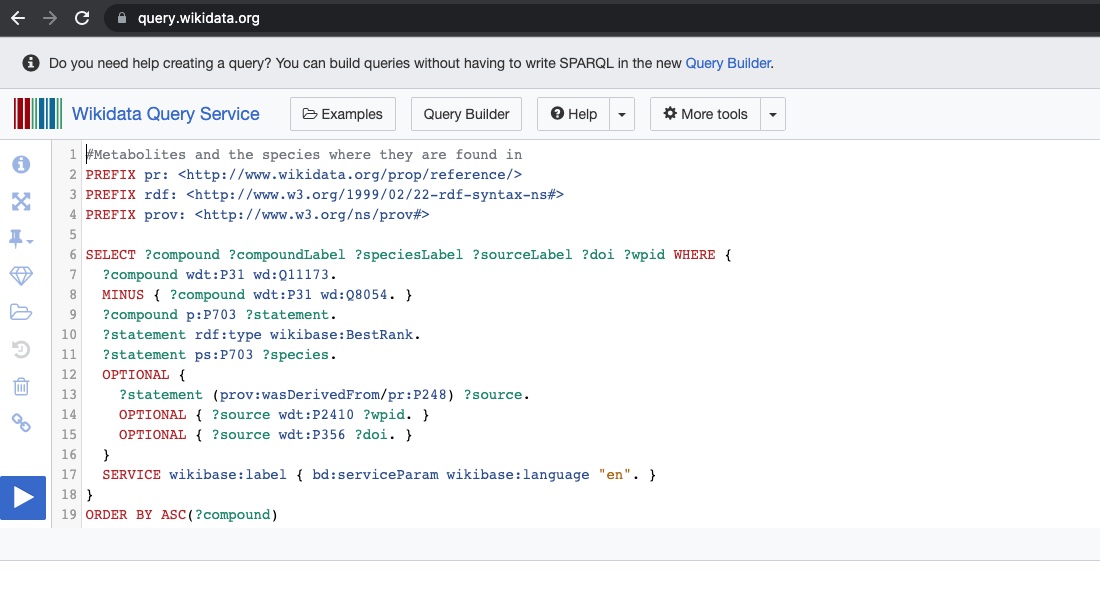
Fig. 9 A SPARQL query over Wikidata RDF endpoint¶
SPARQL query example on WikiData: Cell lines with names that could also be URLs
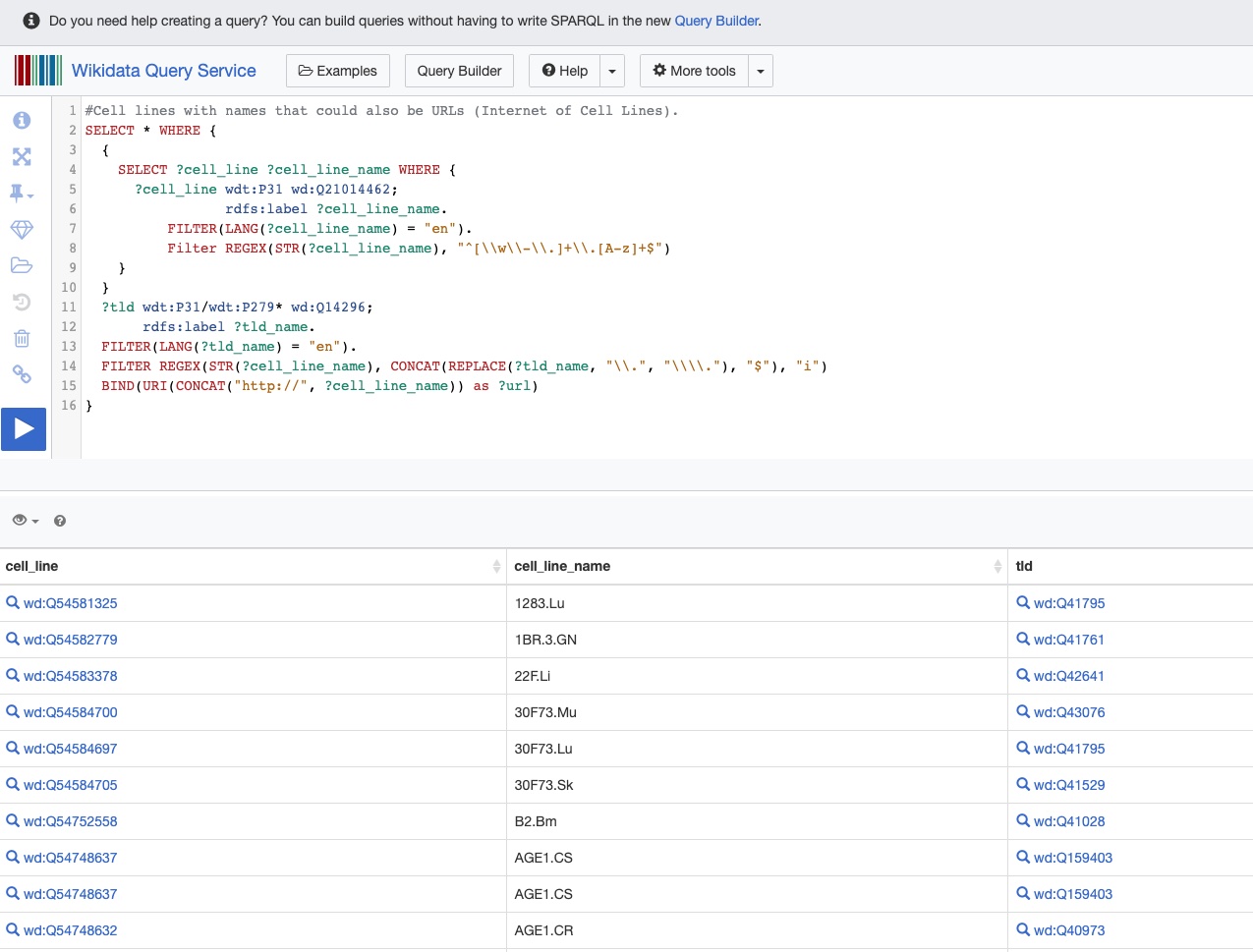
Fig. 10 A SPARQL query over Wikidata RDF endpoint: Cell lines with names that could also be URLs (Internet of Cell Lines)¶
Wikidata provides a list of SPARQL query examples, see here.
2. Property graphs¶
Like RDF graphs, property graphs(PG) are also used to build knowledge graphs.
However, unlike RDF graphs, property graphs allows the vertices (aka the edge) to carry annotations,
which can be queried. These annotations are called properties hence the denomination Property graphs,
also referred to as Labeled Property Graphs or LPG.
Property graphs have inherent interesting properties which set them aside from RDF graphs.
For instance, in PG/LPG, both nodes and edges have:
(1..1) identifier(1..n) annotations, in the form of sets of{key:value}pairs
In addition, edges have a Type.
Important
The main thing to remember is that what distinguishes RDF graphs from Labeled Property graphs is that fact that the edges in a LPG can carry annotations. This allows for interesting properties which affect the querying options available to developers and data miners. It is this difference that has been exploited to develop an entire environment of software solutions supported LPG based knowledge representation and exploitation.
These property graphs constitute the main data structure supported by two key resources which have gain notoriety
and visibility: one open-source and one commercial. In the following sections, we will provide a brief summary of their
salient points and also, limitations.
2.1 Apache Tinkerpop framework and Gremlin graph traversal language¶
The Apache foundation released in 2009 the Apache Tinkerpop framework as an open source initiative, licensed under Apache License 2.0 terms.
Apache Tinkerpop is a vendor agnostic graph database storage framework using on
property graphsas underlying data structure. As a framework, Tinkerpop is developed as an abstract layer, meant to ensure vendor neutrality, allowing developers to decide about architecture choices and tests while coding against a standardized interface.Apache Gremlin is the query language powering Tinkerpop and its interactions with the underlying data stored as property graphs. Gremlin is a
graph traversal languageoptimized for speed and fast access. Defined by Marko Rodrigues, Gremlin language specifications are detailed in the following publication 7 and is supported by libraries available in the most popular programming language. Extensive documentation and training is available from the dedicated Apache project pages, including a set ofrecipesto make the most of the Gremlin language.
Below is an example of a Gremlin instruction as coded in python:
from gremlin_python.process.traversal import T
from gremlin_python.process.traversal import Cardinality
id = T.id
single = Cardinality.single
def create_vertex(vid, vlabel):
# default database cardinality is used when Cardinality argument is not specified
g.addV(vlabel).property(id, vid). \
property(single, 'name', 'Apache'). \
property('lastname', 'Tinkerpop'). \
next()
A Gremlin query to answer a request such Get the names of the people vertex "1" knows who are over the age of 30.
g.V()
g.V(1)
g.V(1).values('name')
g.V(1).outE('knows')
g.V(1).outE('knows').inV().values('name')
g.V(1).out('knows').values('name')
g.V(1).out('knows').has('age', gt(30)).values('name')
2.2 Neo4j property graph database and Cypher Query Language¶
Neo4j is a commercial offering for building knowledge graphs relying on property graphs.
Backing the approach, the team developed a dedicated query language for Labeled property graphs.
This language is known as CYPHER.
Neo4j databases have shown promises in biology and bioinformatics for its ability to allow for fast graph traversals,
which matches the requirements of cell biologists, modelers, and computational scientists who need to explore a growing
ensemble of molecular pathways, that is to say graphs of interactions and reactions.
An illustration of that natural fit is the uptake of the Neo4j technology by a project such as Disease Maps,
Reactome 5 and KnetMiners 4.
The Reactome database builds on Neo4j to allow navigation of reactions and pathways.
A complete tutorial to query Reactome using the Cypher language to interrogate the underlying Neo4j store is
available here.
//All reactions for the pathway with stable identifier R-HSA-198933
MATCH (p:Pathway{stId:"R-HSA-983169"})-[:hasEvent*]->(rle:ReactionLikeEvent)
RETURN p.stId AS Pathway, rle.stId AS Reaction, rle.displayName AS ReactionName
CYPHER query example on Reactome: Comparison with SQL
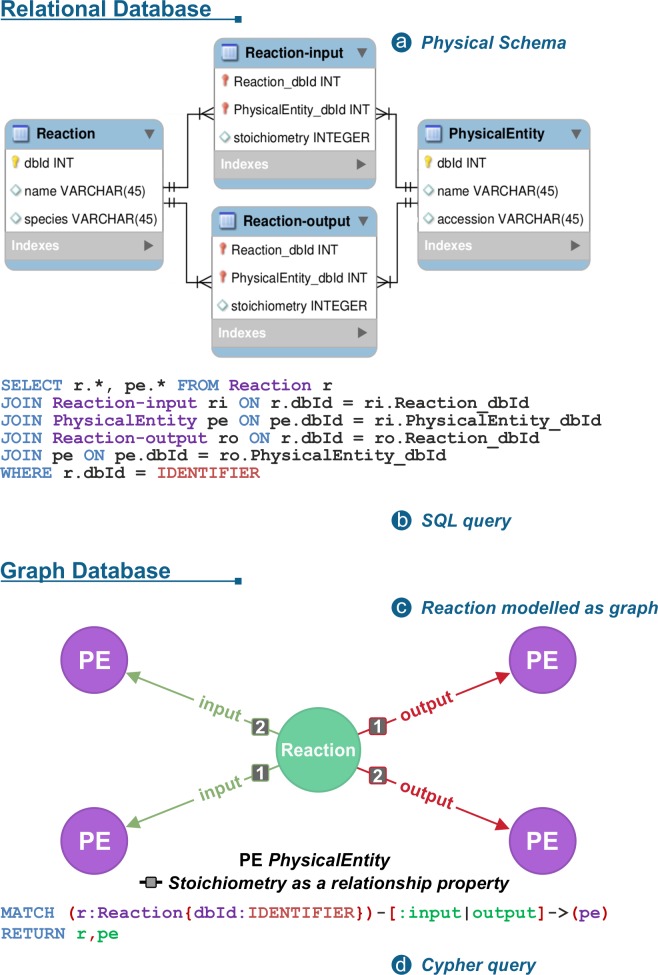
Fig. 11 CYPHER query example on Reactome: Comparison with SQL. From Fabregat et al,2018. 10.1371/journal.pcbi.1005968¶
The [PDBe-KB](PDBe-KG https://www.ebi.ac.uk/pdbe/pdbe-kb/graph-download) is another relevant resources in the fields of bioinformatics which is available as a Neo4j graph database. PDBe-KB is a community-driven resource managed by the PDBe team, collating functional annotations and predictions for structure data in the PDB archive, the content of which is distributed under CC-BY-4 license. PDFe-KB can be downloaded [here](PDBe-KG https://www.ebi.ac.uk/pdbe/pdbe-kb/graph-download).
While Neo4j origin dates back to 2000, the company recently grabbed headlines for a very successful IPO which netted circa 350 millions USD of venture capital funding. Such a success is testament to the robustness and validity of the approach taken by the group of computer scientists who built the solutions from the ground up to deliver one of the most successful solution for property graph based knowledge representation and querying making full use of Neo4j representation and infrastructure capabilities.
Neo4J open-sourced the Cypher query language and made it available via GitHub as part of the open-cypher project
A large body of documentation and training material is available, supporting developers in getting up to speed with the language.
More recently, Neo4j also joined the graphQL foundation, “a neutral, non-profit home for the GraphQL assets and ongoing collaboration, and hosted by The Linux Foundation” to quote the landing page of the organization.
Why this matters?
It matters because GraphQL specification allows building robust Application Programming Interface and make it easier to build full stack applications from scratch while offering great flexibility in interfacing with the data. But the main idea here is that the
GraphQLway of representing data is a natural fit to database systems which are nativelygraph databases, such as Neo4j.
3. RDF graphs or Labeled Property graphs, which has the upper hand?¶
Sometimes, nothing beats a diagram to drive forward the strengths and the weaknesses of technical stacks. In the diagram presented in figure 4, which provides an overview of the specifications and standards available to support the knowledge graph, two things strike the keen reader:
RDF graphs technology is backed by a large number of W3C standards for the semantic web.
W3C RDF
WC3 RDFS
W3C OWL
W3C SHACL
W3C SPARQL 1.1
Content Validation
RDF graphs content can be validated (i.e. checked) with 2 equally functional
constraintlanguages, SHEX and SHACL. Using these languages, the “shape” of the RDF graph can be controlled.With property graphs, until the introduction of an adapter in the Neo4j library, this task proved hard to achieve. This becomes a drawback as it means a property graph database may become
contaminatedwith incorrect data.
Besides this point, two additional features can give RDF graphs the edge (no pun intended!) over labeled property graphs (even if it also depends on the use cases and ultimate goal):
IRI support and Linked data
the RDF specification by construction allows the use of IRI to uniquely identify nodes and have globally unique resolvable identifiers for resources.
Native semantics and the possibility of inference
Backed by RDFS and OWL semantics, RDF graphs are built on a technology stack developed to deliver the
the semantic webas envisioned by Sir Tim Berners-Lee. Tools known as reasoners , use first order logic to perform type and instance classification but also generateentailments, which correspond to new statements resulting from inference made from the explicit assertions in the underlying ontologies or graph.
Worth of notice, comparisons between the two approaches do exist. For instance, Alocci and colleagues ran a comparison of between RDF triple stores and Property graph in the context of Glycan substructure and their assessment can serve as basis for deciding to go for on or the other 2.
4. Towards a unified standard for querying LPG: GQL specifications¶
What now?
In the previous 2 sections, we have highlighted 2 of the most successful projects leveraging the power of
Property Graphs as data structure for storing information and building knowledge graphs.
However, none of these implementations are officially an approved W3C standards. While it does not really matter
at this stage, it was realized that such fragmentation was hurting and in 2019, the open Graph Query Language (oGQL)
working group released an initial specification.
Conclusion¶
Knowledge graphs are a powerful and flexible way to represent information. Their properties and features lend themselves to data driven approaches and make them a data structure of choice for certain types of artificial intelligence and machine learning applications. As with all technical solutions, it is no panacea, no silver bullet. They however are a technology which can greatly enhance certain tasks and are of particular relevance for representing metadata, data about the data in a FAIR way.
What to read next:¶
Having read about what knowledge graphs are and their typology, the next key thing is to learn about how are knowledge graphs generated. This is when we need to introduce the notion of semantic models and ontologies and when knowledge engineers step in.
The main concerns are to 1) provide foundational domain models as well as 2) instantiations of data under these models. To take a simple example, let’s consider converting natural text into a data structure which can be easily manipulated by software agents.
`P53 gene transcript` `binds to` `DNA`.In order to do this, several steps are necessary and some requires specific resources to be available. One step is known as ‘Name Entity Recognition’ (NER), which as the name says aims to cast strings into specific bins such ‘noun’, ‘verb’, ‘qualifier’, etc. Another step, following the NER step would be the anchoring of these entities to a semantic framework and this step is known as ‘semantic mark-up’ or sometimes ‘mapping to ontologies’.
Ontologies are the resources which provide this layer of semantics which make it possible to integrate data in knowledge graphs. Ontologies are formal resources developed by experts in a domain. Ontologies also rely on RDF language for their representation but their purpose is different.
These ontologies set the rules and constraints grounded in first order logic to define a type.
The representation is neat and tidy but one can not help noticing that all the strings are stored as free text. A problem arises when trying to accumulate knowledge from different sources. One needs to add a layer of semantics to
typeeach of the element of an RDF statement.@prefix go: <go:> @prefix uniprot: <uniprot:> @prefix chebi: <chebi:> P53 gene transcript a RDF:type go:Transcript; DNA a RDF:type chebi:molecularEntity binds_toThis taster is really meant to encourage our readers to dive into the following contents, which range from how to build controlled terminology, how to select ontologies or how to perform NER. Check out those content and recipes!
Constructing knowledge graphs and their biomedical applications 6, a review by Nicholson & Greene
References¶
References
- 1
W3c sparql query language 1.1. URL: https://www.w3.org/TR/sparql11-overview/.
- 2
D. Alocci, J. Mariethoz, O. Horlacher, J. T. Bolleman, M. P. Campbell, and F. Lisacek. Property Graph vs RDF Triple Store: A Comparison on Glycan Substructure Search. PLoS One, 10(12):e0144578, 2015.
- 3
Tim Berners-Lee, James Hendler, and Ora Lassila. The semantic web. Scientific American, 284(5):34–43, May 2001. URL: http://www.sciam.com/article.cfm?articleID=00048144-10D2-1C70-84A9809EC588EF21.
- 4
M. Brandizi, A. Singh, C. Rawlings, and K. Hassani-Pak. Towards FAIRer Biological Knowledge Networks Using a Hybrid Linked Data and Graph Database Approach. J Integr Bioinform, Aug 2018.
- 5
A. Fabregat, F. Korninger, G. Viteri, K. Sidiropoulos, P. Marin-Garcia, P. Ping, G. Wu, L. Stein, P. D’Eustachio, and H. Hermjakob. Reactome graph database: Efficient access to complex pathway data. PLoS Comput Biol, 14(1):e1005968, 01 2018.
- 6
D. N. Nicholson and C. S. Greene. Constructing knowledge graphs and their biomedical applications. Comput Struct Biotechnol J, 18:1414–1428, 2020.
- 7
Marko A. Rodriguez. The gremlin graph traversal machine and language (invited talk). In Proceedings of the 15th Symposium on Database Programming Languages, DBPL 2015, 1–10. New York, NY, USA, 2015. Association for Computing Machinery. URL: https://doi.org/10.1145/2815072.2815073, doi:10.1145/2815072.2815073.
What to read next?¶

FAIRsharing records appearing in this recipe:
- GitHub
- Protein Data Bank Format (PDB)
- Protein Data Bank in Europe (PDBe)
- Protein Data Bank in Europe - Knowledge Base (PDBe-KB)
- Reactome
- Resource Description Framework (RDF)
- Resource Description Framework Schema (RDFS)
- Shapes Constraint Language (SHACL) (SHACL)
- Simple Protocol and RDF Query Language Overview (SPARQL)
- The FAIR Principles (FAIR)
- Web Ontology Language (OWL)
- Wikidata
Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
University of Oxford |
Writing - Original Draft, Editing, Conceptualization |
||||
University of Oxford |
Writing - Review & Editing |
||||
University of Oxford |
Writing - Review & Editing |
||||
EMBL-EBI |
|
Review & Editing |
|||
Fraunhofer Institute |
Review & Editing |


