Public knowledge graph resources for life sciences¶

Main Objectives¶
In the following sections, we will cover the following topics:
the known reference knowledge graphs to integrate with.
the different scientific contexts.
the impact for Pharma industry and EFPIA more broadly.
Introduction¶
When it comes to research carried out by academic and industry ventures such as the Innovative Medicine Initiative of these European Union, three main scenarios can be outlined:
Identification of new drug targets, as it the case for the IMI Resolute project for instance, which focuses on characterizing solute carriers and identifying potential candidates for “druggability”.
Development of new therapeutic products and techniques given a specific disease area, as in the IMI AETIONOMY project.
Discovery of new drugs against pathogens as for instance the New Drugs for Bad Bugs (ND4BB) project.
This gives a backdrop against which work to enhance interoperability of data generated by these projects needs to be aware of the existing data landscape.
The purpose of the present content type is to provide a basic survey of the existing, and the relevance to specific research contexts and tasks. This information may be useful when deciding on particular technical aspects such as choice of data interface, choice of terminology and controlled terminologies or, choice of data formats.
We therefore focus the dataset scan on main entities such as Drugs / Chemicals and Disease / Clinical Research Data. Such datasets are popularly known as Knowledge Graphs (KGs).
Additionally, the availability of such information in specialized graph representations was also included as a selection criteria. Therefore, services providing SPARQL endpoint and Graph API for dataset access were considered and the following sections providing specific information and references for each of the resources and datasets are considered
A catalog of SPARQL Endpoints for Life Sciences: Yummydata¶
Yummydata is a catalog of SPARQL endpoints of interest to the biomedical community, maintained by the Database Center for Life Sciences in Kashiwa, Japan 13.
Database URL: http://yummydata.org/
The resource can be viewed as a registry to resources as well as a monitoring services, which regularly checks the “health status” of the resources it indexes.
The “health status” is reported as a “Umaka Score” which consolidates endpoint status according to six dimensions: Availability, Freshness, Operation, Usefulness, Validity, and Performance.
It should be noted that Yummydata does not differentiate between semantic artefacts, such as an ontology, or a dataset represented in RDFLinked data.
The authors of the resource highlight this point and underline that being able to distinguish between these types of graph explicitly may be required in the future.
YummyData
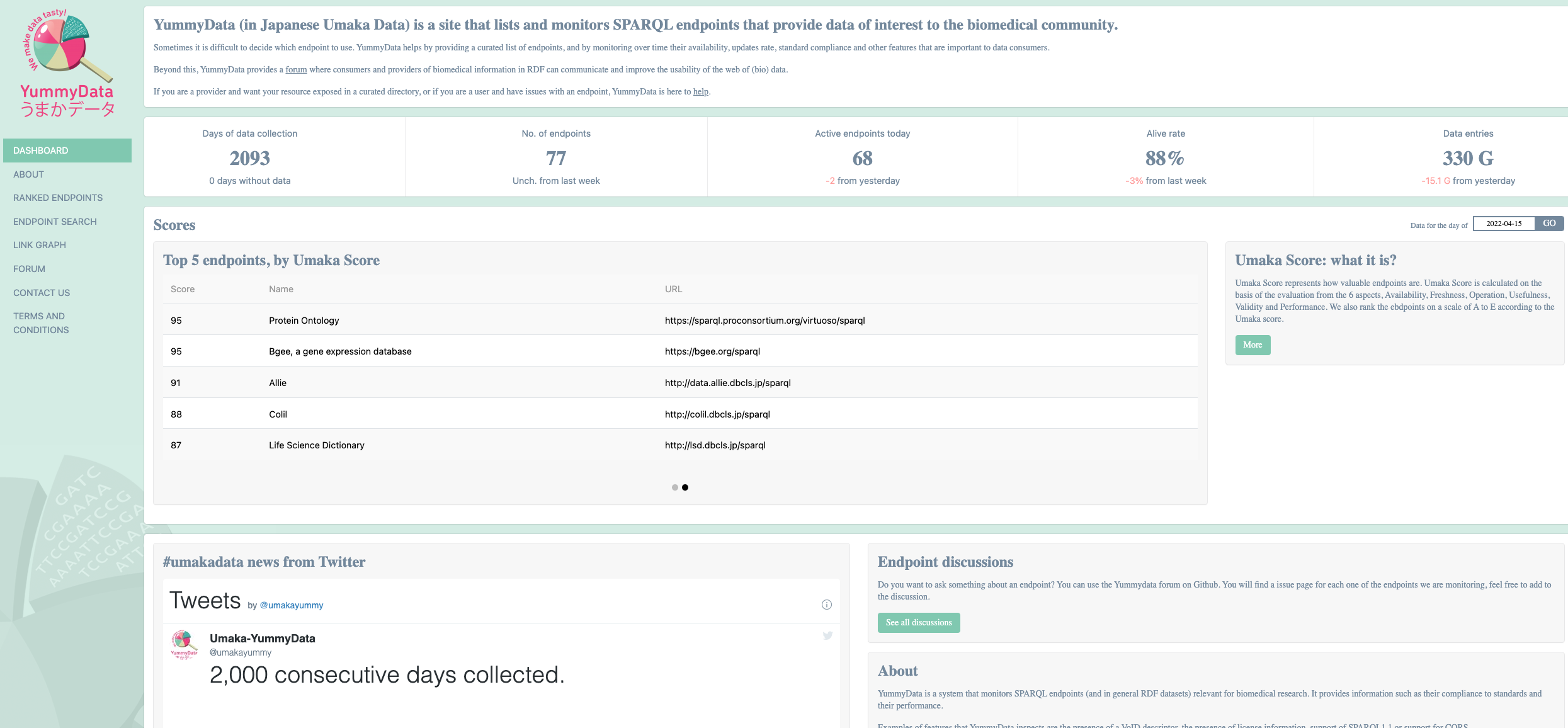
Fig. 13 YummyData, a registry of sparql endpoints¶
Resources for Druggable Targets Predictions¶
1. OpenTarget¶
The OpenTarget project is a public private partnership aiming at providing a central resource based on public domain data to enable target identification and prioritisation, based on functional genomics data and GWAS data 10.
Using a genome wide, evidence based approach, the OpenTarget project offers the possibility to obtain reliable information about validated targets against which design active compounds. Such approach has been shown to yield higher success in progressing potential therapeutic compounds along the clinical trial pipeline.
All the data generated by the OpenTarget project is available for download and is accessible via
a GraphQL application programming interface. https://api.genetics.opentargets.org/graphql
a Google Big Query interface. https://platform-docs.opentargets.org/data-access/google-bigquery
a ftp download. https://platform.opentargets.org/downloads/data
More recently, the dataset is also available from Amazon Web Services
OpenTarget project in AWS

Fig. 14 OpenTarget project in AWS¶
The associated code is also open sourced, under an Apache License 2.0. More information is available here: https://genetics-docs.opentargets.org/data-access/data-download
GitHub Repository: https://github.com/opentargets
The OpenTarget project integrates data from several key resources listed in the project’s main publication. From an integration scenario standpoint, it is worth noting that data are annotated with semantic resources such as:
EFO: Experimental Factor Ontology and associated open target slim EFO-OTAR
MONDO: Diseases
HPO: clinical signs, symptoms and phenotypes
ECO: evidence code ontology to annotate confidence and reliability of findings
query genePrioritisationUsingL2G {
studyLocus2GeneTable(studyId: "FINNGEN_R5_E4_DM2", variantId: "11_72752390_G_A") {
rows {
gene {
symbol
id
}
yProbaModel
yProbaDistance
yProbaInteraction
yProbaMolecularQTL
yProbaPathogenicity
hasColoc
distanceToLocus
}
}
}
#!/usr/bin/env python3
# Import relevant libraries for HTTP request and JSON formatting
import requests
import json
# Set study_id and variant_id variables
study_id = "FINNGEN_R5_E4_DM2"
variant_id = "11_72752390_G_A"
# Build query string
query_string = """
query genePrioritisationUsingL2G($myVariantId: String!, $myStudyId: String! ){
studyLocus2GeneTable(studyId: $myStudyId, variantId: $myVariantId){
rows {
gene {
symbol
id
}
yProbaModel
yProbaDistance
yProbaInteraction
yProbaMolecularQTL
yProbaPathogenicity
hasColoc
distanceToLocus
}
}
}
"""
# Set variables object of arguments to be passed to endpoint
variables = {"myVariantId": variant_id, "myStudyId": study_id}
# Set base URL of Genetics Portal GraphQL API endpoint
base_url = "https://api.genetics.opentargets.org/graphql"
# Perform POST request and check status code of response
r = requests.post(base_url, json={"query": query_string, "variables": variables})
print(r.status_code)
# Transform API response into JSON
api_response_as_json = json.loads(r.text)
# Print first element of JSON response data
print(api_response_as_json["data"]["studyLocus2GeneTable"]["rows"][0])
2. Pharos¶
Pharos is a US NIH funded effort, which can be viewed as the counterpart to the European lead OpenTarget project 9.
Pharos comes on the back of initiative such as Illuminating the Druggable Genome (IDG) program (https://commonfund.nih.gov/idg/index) and Target Central Resource Database (TCRD).
Pharos integrates data from 66 different data sources, the list of which is available here
Pharos information is organized around:
Diseases
Targets
Ligands
Pharos
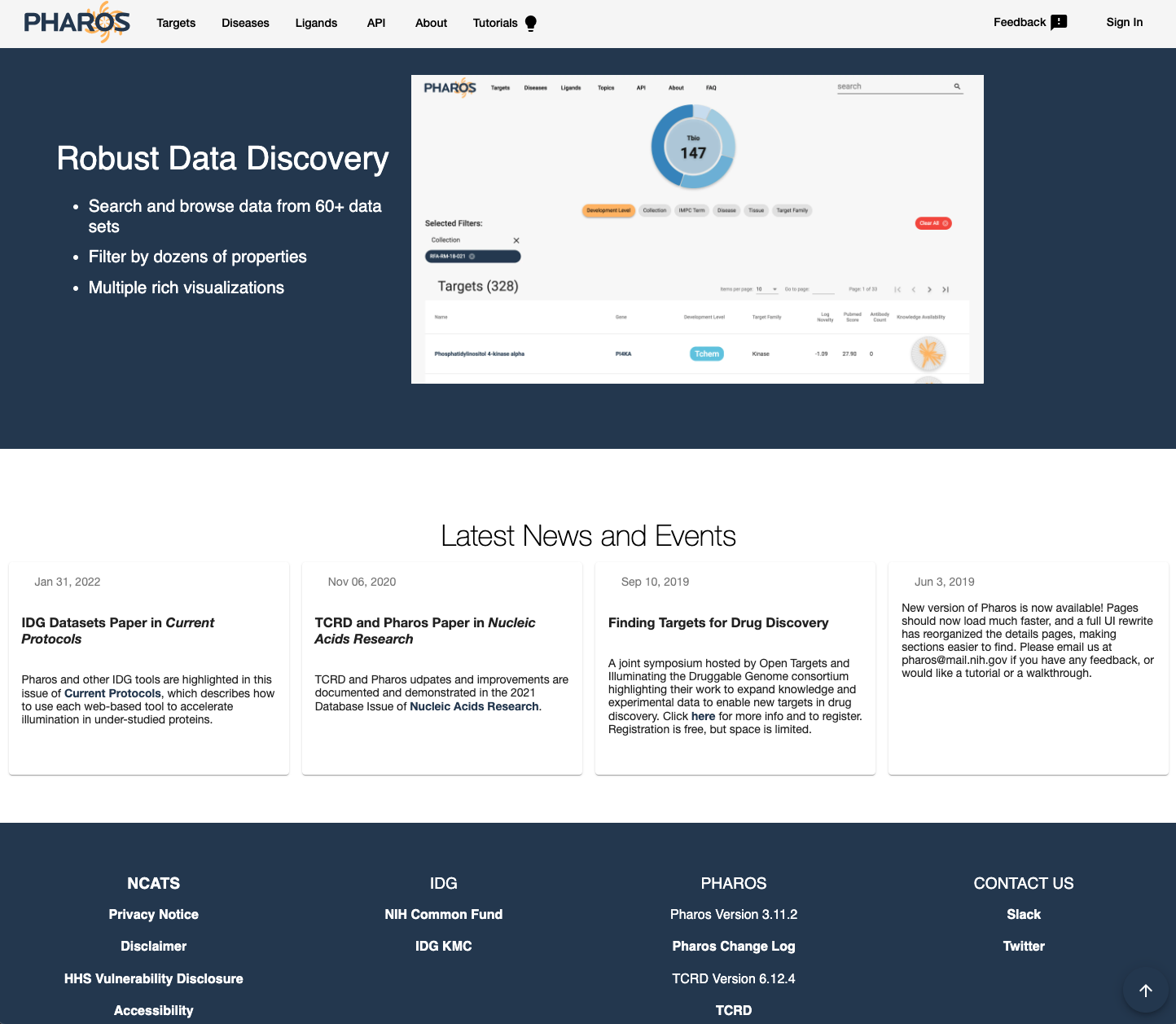
Fig. 15 Pharos¶
All the Pharos data can be queried via a GraphQL API: https://pharos.nih.gov/api
From an integration scenario standpoint, Pharos integrates data with custom-built ontology, the Drug Target Ontology (DTO).
The DTO reuses terms from well established resources such as:
DO: Disease Ontology
GO: Gene Ontology
BAO: BioAssay Ontology
GPCRO: GPCR Ontology
Pharos source code is available under the MIT license, from https://spotlite.nih.gov/ncats/pharos.
3. Hetionet: an integrative network of disease¶
The main purpose of Hetionet was to predict new uses for existing drugs, as part of the “Project Rephetio” .
Hetionet Version 1.0 corresponds to an ‘heterogeneous network’ (or HetNet) and contains 47,031 nodes of 11 types and 2,250,197 relationships of 24 types, integrating data from 29 Resources to connect compounds, diseases, genes, anatomies, pathways, biological processes, molecular functions, cellular components, pharmacologic classes, side effects, and symptoms.
For more details about the full list of source, consult the publication 8.
The network is available in four formats:
JSON — see
hetnet/jsonNeo4j — see
hetnet/neo4jannd https://neo4j.het.io/browser/TSV — see
hetnet/tsvmatrix — see
hetnet/matrix
GitHb Repository: https://github.com/hetio/hetionet
license: CC0
From an integration scenario standpoint, Hetionet integrates knowledge semantically by anchoring entities to several ontology “slims”, which are simplified versions of larger semantic artefacts.
DO: disease ontology slim, with 137 terms considered.
MESH: for signs and symptoms, with 438 terms considered
SIDER: for side effects
GO: Biological processes, cellular components, and molecular functions.
UBERON: for anatomical terms.
Mappings provide connection to chemical information via the use of InChI keys to unambiguously specific chemical structures.
4. Drug Repurposing Knowledge graph¶
Drug Repurposing Knowledge Graph (DRKG) is a comprehensive biological KG relating genes, compounds, diseases, biological processes, side effects, and symptoms.
DRKG includes information from six existing databases including DrugBank, Hetionet, GNBR, String, IntAct and DGIdb, and data collected from recent publications particularly related to Covid-19.
It includes 97,238 entities belonging to 13 entity-types; and 5,874,261 triples belonging to 107 edge-types.
GitHb Repository: https://github.com/gnn4dr/DRKG/
A number of jupyter notebooks are available from the repository. They show how to use the KGs to explore drug repurposing possibilities with help of a pretrained model:
Knowledge Graph training notebook: https://github.com/gnn4dr/DRKG/blob/master/drug_repurpose/COVID-19_drug_repurposing_via_genes.ipynb
5. OpenBiolink¶
OpenBioLink is an open benchmark dataset that serve as training or evaluation datasets for link prediction models 6, 11. It is an ensembl of large independent resource connecting drugs to disease via proteins. These resources include STRING, GO, DisGeNet, HPO, STITCH, CTD, HPO, DrugCentral, and SIDER.
Taking a deeper dive into the KG statistics, there are 24,806 entities that are either drugs, proteins, indications or phenotype and 2,35.881 triples that are part of 4 edge-types.
GitHb Repository: https://openbiolink.github.io/
from openbiolink.obl2021 import OBL2021Dataset
dl = OBL2021Dataset()
train = dl.training # torch.tensor of shape(num_train,3)
valid = dl.validation # torch.tensor of shape(num_val,3)
6. Clinical Knowledge Graph¶
The Clinical Knowledge Graph is an open source platform currently comprised of more than 16 million nodes and 220 million relationships to represent relevant experimental data, public databases and the literature 12.
Documentation: https://ckg.readthedocs.io/en/latest/INTRO.html
From an integration scenario standpoint, the CKG integrates data using the following semantic resources:
DO: Disease Ontology
BTO: Brenda Tissue Ontology for anatomical entities and tissues.
EFO: Experimental Factor Ontology
HPO: Human Phenotype Ontology
GO: Gene Ontology
SNOMED-CT: which defines clinical terms and their associative relationships.
The code for CKG is available from GitHub at: https://github.com/MannLabs/CKG under a MIT License.
CKG can be loaded and queried via a Neo4j graph database, a data dump of which is available from: https://data.mendeley.com/datasets/mrcf7f4tc2/1.
7. AstraZeneca Biological Insights Knowledge Graph BIKG¶
The BIKG is a resource developed by AstraZeneca to combine both public and internal data to facilitate knowledge discovery using machine learning over KG.
To build it, AstraZeneca developers created dedicated information extraction pipeline for mining unstructured text such as full text publications using Natural Language Processing (NLP) techniques, as well as information obtained from specialized databases such as ChEMBL or Ensembl for chemical and gene products respectively, but also other datasets such as Hetionet and Open Target.
The latest version of the BIKG consists of 10.9m nodes (of 22 types) and over 118 million unique edges (of 59 types, forming 398 different triples)” 7.
Interestingly, the BIKG pipelines also allow third-party datasets to be integrated. This step involves an ETL into the BIKG semantic model and the BIKG upper ontology (BIKG ULO).
For integration purpose, the BIKG ULO is compatible with ontologies such as:
UBERON: for anatomical terms
GO: Gene Ontology for Biological processes, cellular components, and molecular functions.
BIKG can be accessed via a GraphQL API and the underlying data can be exported in 2 different graph formats, RDF or Neo4j/CSV for linked data graph or property graph representations respectively.
Interestingly, two use cases for using the BIKG are discussed in the associated publications and clearly outline FAIRification goals. These use cases are:
Target Identifications
CRISPR Screen Hit ranking
PyKEEN¶
PyKEEN (Python KnowlEdge EmbeddiNgs) is a Python package designed to train and evaluate knowledge graph embedding models (KGEM) (incorporating multi-modal information) 1, 2.
GitHub Repository: https://github.com/pykeen/pykeen
So, as such, PyKEEN is more a tool than a KG, however PyKEEN contains many reference KGs and datasets which can be used for a variety of data science tasks and machine learning applications. PyKEEN is mainly geared towards helping developers train KGEM for tasks such as link prediction.
The following figure shows a snippet of the various KGs available from the python package.
PyKEEN datasets
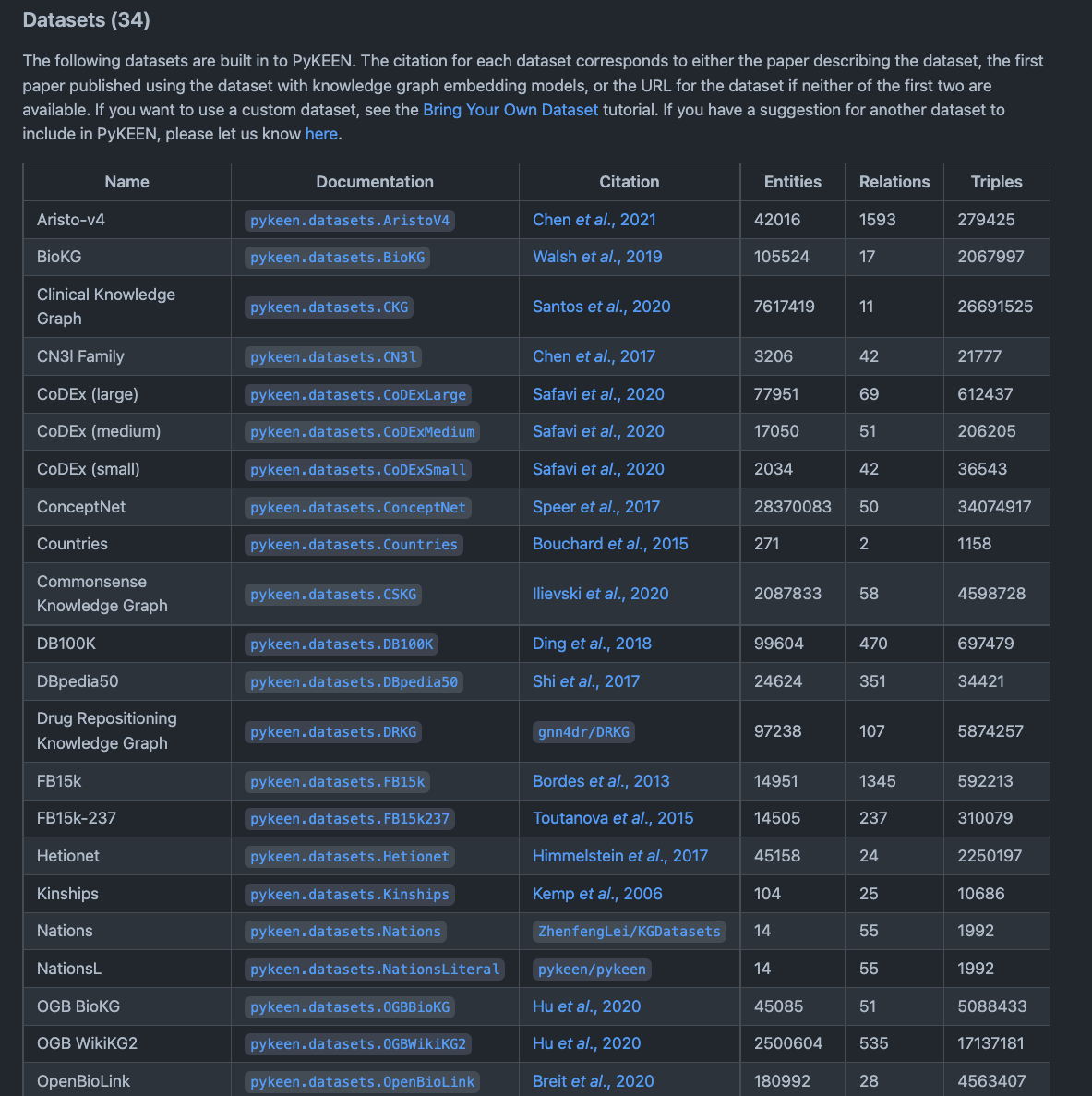
Fig. 16 A screenshot of the README on the PyKEEN GitHub repository¶
pykeen
This is one of the main reason why we mention this library as it provides a context to data integration scenarios along with the datasets and tooling to do so.
from pykeen.pipeline import pipeline
result = pipeline(
model='TransE',
dataset='nations',
)
To see an example of how to use the library, we highly recommended reading the Medium blog post and consult the associated jupyter notebook, which shows how to use your own dataset, understand the evaluation, and make novel link predictions:
Link Prediction example notebook: https://github.com/tomasonjo/blogs/blob/master/pykeen/Hetionet - RotatE.ipynb
Conclusion¶
With this recipe, the intent was to raise awareness in the wealth of Life Science information available as Knowledge Graph and ongoing integrative efforts. We have also tried to convey information about the license for each of the resources as well as some insights into the curation protocols specific to each the KG. Finally, we wanted to highlight how some KGs are already used to train models and make predictions in the context of drug repurposing for instance. This is of particular interest in the context of the Innovative Medicine Initiative but not only, obviously. It certainly should provide food for thoughts when engaging with a FAIRification process and should help define FAIRification goals as well as the process to perform in order to be able to be compatible with specific resources for a specific task at hand.
What to read next?¶
Implications of Topological Imbalance for Representation Learning on Biomedical Knowledge Graphs 4
A Review of Biomedical Datasets Relating to Drug Discovery: A Knowledge Graph Perspective 3

FAIRsharing records appearing in this recipe:
- BRENDA tissue / enzyme source (BTO)
- BioAssay Ontology (BAO)
- ChEMBL
- Comma-separated Values (CSV)
- Comparative Toxicogenomics Database (CTD)
- DisGeNET
- Disease Ontology (DOID)
- Drug Target Ontology (DTO)
- DrugBank
- DrugCentral
- Ensembl
- Evidence and Conclusion Ontology (ECO)
- Experimental Factor Ontology (EFO)
- Gene Ontology (GO)
- GitHub
- Human Phenotype Ontology (HP)
- IUPAC International Chemical Identifier (InChI)
- IntAct molecular interaction database (IntAct)
- JavaScript Object Notation (JSON)
- Medical Subject Headings (MESH)
- Mendeley Data
- Minimum Information About GWAS Experiment (GWAS)
- Monarch Disease Ontology (MONDO)
- Open Targets
- STRING
- Search Tool for Interactions of Chemicals (STITCH)
- Side Effect Resource (SIDER)
- Simple Protocol and RDF Query Language Overview (SPARQL)
- Tab-separated values (TSV)
- Target Central Resource Database (TCRD)
- UBER anatomy ONtology (UBERON)
- Uniform Resource Locator (URL)
References¶
Reference
- 1
M. Ali, M. Berrendorf, C. T. Hoyt, L. Vermue, M. Galkin, S. Sharifzadeh, A. Fischer, V. Tresp, and J. Lehmann. Bringing Light Into the Dark: A Large-scale Evaluation of Knowledge Graph Embedding Models under a Unified Framework. IEEE Trans Pattern Anal Mach Intell, Nov 2021.
- 2
M. Ali, C. T. Hoyt, D. Domingo-Fernández, J. Lehmann, and H. Jabeen. BioKEEN: a library for learning and evaluating biological knowledge graph embeddings. Bioinformatics, 35(18):3538–3540, 09 2019.
- 3
Stephen Bonner, Ian P Barrett, Cheng Ye, Rowan Swiers, Ola Engkvist, Andreas Bender, Charles Tapley Hoyt, and William L Hamilton. A review of biomedical datasets relating to drug discovery: a knowledge graph perspective. 2021. URL: https://arxiv.org/abs/2102.10062, doi:10.48550/ARXIV.2102.10062.
- 4
Stephen Bonner, Ufuk Kirik, Ola Engkvist, Jian Tang, and Ian P. Barrett. Implications of topological imbalance for representation learning on biomedical knowledge graphs. CoRR, 2021. URL: https://arxiv.org/abs/2112.06567, arXiv:2112.06567.
- 5
A. Breit, S. Ott, A. Agibetov, and M. Samwald. OpenBioLink: a benchmarking framework for large-scale biomedical link prediction. Bioinformatics, 36(13):4097–4098, 07 2020.
- 6
Daniel Domingo-Fernández, Yojana Gadiya, Abhishek Patel, Sarah Mubeen, Daniel Rivas-Barragan, Chris W. Diana, Biswapriya B. Misra, David Healey, Joe Rokicki, and Viswa Colluru. Causal reasoning over knowledge graphs leveraging drug-perturbed and disease-specific transcriptomic signatures for drug discovery. bioRxiv, 2021. URL: https://www.biorxiv.org/content/early/2021/09/15/2021.09.12.459579, arXiv:https://www.biorxiv.org/content/early/2021/09/15/2021.09.12.459579.full.pdf, doi:10.1101/2021.09.12.459579.
- 7
David Geleta, Andriy Nikolov, Gavin Edwards, Anna Gogleva, Richard Jackson, Erik Jansson, Andrej Lamov, Sebastian Nilsson, Marina Pettersson, Vladimir Poroshin, Benedek Rozemberczki, Timothy Scrivener, Michael Ughetto, and Eliseo Papa. Biological insights knowledge graph: an integrated knowledge graph to support drug development. bioRxiv, 2021. URL: https://www.biorxiv.org/content/early/2021/11/01/2021.10.28.466262, arXiv:https://www.biorxiv.org/content/early/2021/11/01/2021.10.28.466262.full.pdf, doi:10.1101/2021.10.28.466262.
- 8
D. S. Himmelstein, A. Lizee, C. Hessler, L. Brueggeman, S. L. Chen, D. Hadley, A. Green, P. Khankhanian, and S. E. Baranzini. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. Elife, 09 2017.
- 9
Dac-Trung Nguyen, Stephen Mathias, Cristian Bologa, Soren Brunak, Nicolas Fernandez, Anna Gaulton, Anne Hersey, Jayme Holmes, Lars Juhl Jensen, Anneli Karlsson, Guixia Liu, Avi Ma’ayan, Geetha Mandava, Subramani Mani, Saurabh Mehta, John Overington, Juhee Patel, Andrew D. Rouillard, Stephan Schürer, Timothy Sheils, Anton Simeonov, Larry A. Sklar, Noel Southall, Oleg Ursu, Dusica Vidovic, Anna Waller, Jeremy Yang, Ajit Jadhav, Tudor I. Oprea, and Rajarshi Guha. Pharos: Collating protein information to shed light on the druggable genome. Nucleic Acids Research, 45(D1):D995–D1002, 11 2016. URL: https://doi.org/10.1093/nar/gkw1072, arXiv:https://academic.oup.com/nar/article-pdf/45/D1/D995/8846748/gkw1072.pdf, doi:10.1093/nar/gkw1072.
- 10
David Ochoa, Andrew Hercules, Miguel Carmona, Daniel Suveges, Asier Gonzalez-Uriarte, Cinzia Malangone, Alfredo Miranda, Luca Fumis, Denise Carvalho-Silva, Michaela Spitzer, Jarrod Baker, Javier Ferrer, Arwa Raies, Olesya Razuvayevskaya, Adam Faulconbridge, Eirini Petsalaki, Prudence Mutowo, Sandra Machlitt-Northen, Gareth Peat, Elaine McAuley, Chuang Kee Ong, Edward Mountjoy, Maya Ghoussaini, Andrea Pierleoni, Eliseo Papa, Miguel Pignatelli, Gautier Koscielny, Mohd Karim, Jeremy Schwartzentruber, David G Hulcoop, Ian Dunham, and Ellen M McDonagh. Open Targets Platform: supporting systematic drug–target identification and prioritisation. Nucleic Acids Research, 49(D1):D1302–D1310, 11 2020. URL: https://doi.org/10.1093/nar/gkaa1027, arXiv:https://academic.oup.com/nar/article-pdf/49/D1/D1302/35363725/gkaa1027.pdf, doi:10.1093/nar/gkaa1027.
- 11
Daniel Rivas-Barragan, Sarah Mubeen, Francesc Guim Bernat, Martin Hofmann-Apitius, and Daniel Domingo-Fernández. Drug2ways: reasoning over causal paths in biological networks for drug discovery. PLOS Computational Biology, 16(12):1–21, 12 2020. URL: https://doi.org/10.1371/journal.pcbi.1008464, doi:10.1371/journal.pcbi.1008464.
- 12
Alberto Santos, Ana R. Colaço, Annelaura B. Nielsen, Lili Niu, Philipp E. Geyer, Fabian Coscia, Nicolai J Wewer Albrechtsen, Filip Mundt, Lars Juhl Jensen, and Matthias Mann. Clinical knowledge graph integrates proteomics data into clinical decision-making. bioRxiv, 2020. URL: https://www.biorxiv.org/content/early/2020/05/10/2020.05.09.084897, arXiv:https://www.biorxiv.org/content/early/2020/05/10/2020.05.09.084897.full.pdf, doi:10.1101/2020.05.09.084897.
- 13
Y. Yamamoto, A. Yamaguchi, and A. Splendiani. YummyData: providing high-quality open life science data. Database (Oxford), 01 2018.
Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
University of Oxford |
Writing - Original Draft |
||||
Fraunhofer Institute |
Writing - Review & Editing |

