4. Introducing the DATS model¶

4.1. Main Objectives¶
The main purpose of this recipe is:
Provide a general introduction to the Data Tag Suite model for representing project, study and dataset metadata
Highlight challenges in implementing the model in a data catalogue
4.4. Model overview¶
Data Tag Suits (DATS) 2 is a data description model designed and produced to describe datasets being ingested in DataMed, a prototype for data discovery developed as part of the bioCADDIE project 1. DATS was co-developed by the Oxford e-Research Centre and the NIH Data Common Big Data to Knowledge (BD2K) initiative, where it was used to uniformly represent metadata across a number of projects, including the Genotype-Tissue Expression project (GTEx) and Trans-Omics for Precision Medicine (TOPMed).
DATS is semantically compatible with the Data Catalog Vocabulary (DCAT), a Resource Description Framework (RDF) vocabulary designed to facilitate interoperability among data catalogues published on the web, as well as schema.org (SDO), which is a community-driven effort with a similar interoperability goal to DCAT but a more general-purpose scope.
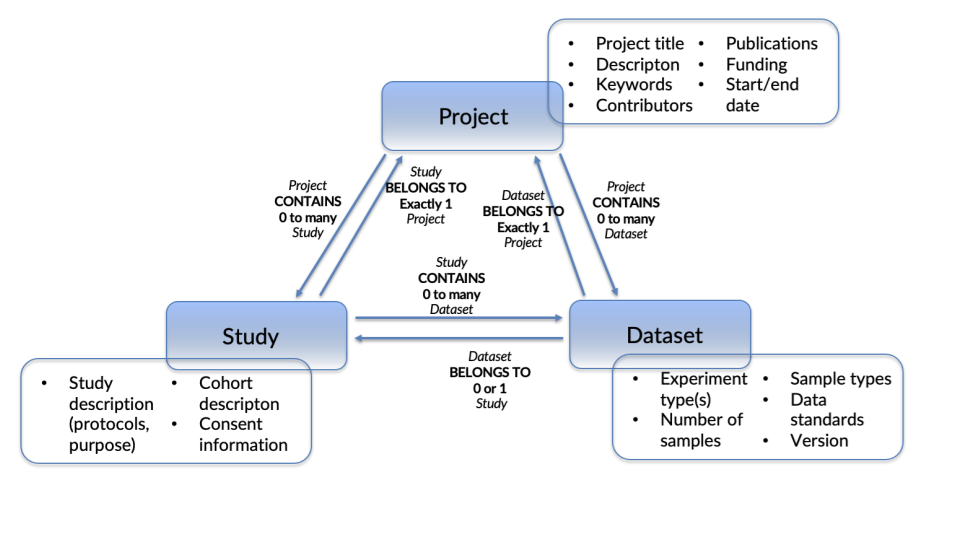
The original DATS model centered around concept of a “Dataset”, an entity that covers technical aspects such as licensing, data types and distributions. The Dataset is produced by or is the input or output of a “Study”, which contains elements that are specific to life, environmental and biomedical science domains, and which models experimental processes, cohorts and protocol information. To meet the requirements of project-generated datasets, the DATS model was extended to include the third core concept of “Project”, covering general information such as title, publications, funding and contributors.
4.4.1. Core objects¶
The DATS model centres around three core objects, “Project”, “Study” and “Dataset”, each of which contains a set of sub-objects, which in turn can be nested down to the lowest unitary object (which contains no further objects), which is the “Annotation”. Project, Study and Dataset relate to each other in a triangular fashion as shown in figure 1.
At first glance, the three core objects make the DATS model reminiscent of the ISA (Investigation, Study, Assay) model for describing experimental processes. There are however several key differences between the two. For one, ISA has a waterfall structure whereby any instance of the model has to contain a Study object between an Investigation and an Assay. In DATS on the other hand, it is possible for a Dataset to be directly dependent on a Project, without any Study information. This is relevant for datasets such as knowledge graphs or synthetic datasets, for which no study metadata may be available. Additionally, DATS focuses on the technical details, availability and use restrictions of datasets whereas the purpose of the ISA model is to describe experimental procedures in great detail. While the DATS Study covers some of this information, it is much less comprehensive or central to the captured data than for ISA.
4.4.1.1. Project¶
The Project object was not part of the original version of DATS. It was added in the second version as a means of linking different studies and datasets under one common parent as well as capturing information such as project contacts or consortium information, publications not linked to one specific study, funding sources, project websites and start/end dates.
4.4.1.2. Study¶
The Study object, although part of the original DATS model, had a less central role in the first iteration than it has in this latest one. Capturing the context in which the Dataset was either generated or used, the Study provides information about the type of study undertaken (e.g. clinical trials or chemical toxicity screens), cohorts (or “studyGroups), input materials, selection criteria and the steps involved in the study.
4.4.1.3. Dataset¶
The Dataset object was the central concept of the initial DATS model but became an equal part in the Project-Study-Dataset triangle in version 2. The object captures technical information about the dataset, such as file types, data standards, versions and licensing as well as links to the actual data if available. It also models information related to the creation of the dataset such as input materials, diseases, biological entities and other similar objects, as well as the types of experiments from which the dataset was derived and any associated dimensions or components.
4.4.2. Sub-objects¶
Each of the three top-level objects references a range of sub-objects, which in turn contain further sub-objects, down to the lowest unitary object (which contains no further objects), which is the “Annotation”. An “Annotation” consists of just two key-value pairs, the “value” and, optionally, the “valueIRI”, designed to capture the Internationalized Resource Identifier (IRI) of an ontology term contextualising the free text “value”. Due to this nested object structure, DATS can be quite opaque to parse for the human reader but allows for easier programmatic processing of the objects. A full overview of the DATS schema can be found on the DataTagSuite Github repository.
4.4.3. Ontology annotations¶
The provision for ontology annotations is at the core of the DATS model, with the smallest object available in the model being the Annotation schema, which simply consists of two key-value pairs: value, which can contain a text string or number (to allow for coded values), and valueIRI, which contains a URI-formatted string representing a concept or ontology term associated with the value. It should be noted that the Annotation object has no required properties, so the valueIRI in particular can be empty.
The model also provides an extension mechanism through a schema object called CategoryValuesPair. This allows the addition of extra properties to the entities in cases where there are no specific properties to deal with the desired property. In order to capture the semantics of the category, CategoryValuesPairs capture both a category as a free-text string and a categoryIRI for the URI of an associated ontology context. The values are of type Annotation and can therefore also have ontology annotations.
DATS does not prescribe the use of specific ontologies in relation to various properties, although it would be advisable to include this type of restriction in a given implementation of the model in order to simply the interoperability between different objects of the same type.
4.4.4. Encoding in JSON-LD¶
DATS objects are encoded in JSON-LD, a method for encoding linked data in the open standard file and data interchange format JSON. JSON-LD is designed around the concept of a “context” to provide additional mappings from JSON to an RDF model. DATS provides three sets of context mappings, to DCAT, schema.org (SDO) and the OBO Foundry Ontologies. None of the three sets of contexts individually cover all properties in the model but they can be used in combination to maximise the semantic linking of the model, likely to increase interoperability, for instance with DCAT based catalogues.
4.5. Implementing DATS in a data catalogue¶
We implemented DATS v2.0 in the IMI Translational Data Catalog. The DATS model’s flexibility presents both a benefit and a challenge in that some concepts could be encoded in a number of ways. Experimental materials for example can be encoded at both the Study level, in Study.input or at the Dataset level, in Dataset.isAbout. In order to encode metadata consistently and simplify indexing and display processes for the IMI Data Catalog, we therefore made a number of assumptions about how to encode certain metadata entities in the model.
4.5.1. Assumptions¶
Study and Dataset objects can’t exist without a Project. This rule was necessary to provide a root for our indexing service.
The Study object encodes information about the input materials to the experiment, such as study subjects in the case of clinical trials or biological molecules in the case of chemical assays. At this level, we collect cohort descriptions, sample sizes, sample sources, types of samples and diseases.
The Dataset object encodes information about the dataset, including technical information such as version number, creation and update dates, data standards and file types. The dataset also lists the experiment types that generated the data, the number and types of samples that contributed to the dataset and any diseases in the samples. These properties can be distinct from the ones at Study level as not all samples collected in a Study may be used to generate a given dataset, e.g. blood samples might be used for sequencing experiments, generating a sequencing dataset, while other tissue samples are used in imaging assays, generating an imaging dataset. The number of samples collected can be different from the cohort sample size (i.e. number of subject), either because multiple samples were collected from each subject or because some samples were excluded from the dataset for quality reasons.
In the implementation of the model, preference as given to specific ontologies in certain contexts, eg MONDO for diseases, UBERON for anatomical components, NCIt, EFO and OBI for experiment types, UO for units etc.
4.6. Conclusion¶
DATS is a flexible data model aimed at biomedical datasets.
DATS utilises the power of JSON-LD to encode semantics at all levels of the model.
DATS’ flexibility does mean that consistent implementation of the model in a specific context requires assumptions to be made.
4.6.1. What should I read next?¶
4.7. References:¶
References
- 1
L. Ohno-Machado, S. A. Sansone, G. Alter, I. Fore, J. Grethe, H. Xu, A. Gonzalez-Beltran, P. Rocca-Serra, A. E. Gururaj, E. Bell, E. Soysal, N. Zong, and H. E. Kim. Finding useful data across multiple biomedical data repositories using DataMed. Nat Genet, 49(6):816–819, 05 2017.
- 2
S. A. Sansone, A. Gonzalez-Beltran, P. Rocca-Serra, G. Alter, J. S. Grethe, H. Xu, I. M. Fore, J. Lyle, A. E. Gururaj, X. Chen, H. E. Kim, N. Zong, Y. Li, R. Liu, I. B. Ozyurt, and L. Ohno-Machado. DATS, the data tag suite to enable discoverability of datasets. Sci Data, 4:170059, 06 2017.

Learn more about:

FAIRsharing records appearing in this recipe:
- DatA Tag Suite (DATS)
- Data Catalog Vocabulary (DCAT)
- Experimental Factor Ontology (EFO)
- JavaScript Object Notation (JSON)
- JavaScript Object Notation for Linking Data (JSON-LD)
- Monarch Disease Ontology (MONDO)
- NCI Thesaurus (NCIt)
- OBO Foundry (OBO)
- Ontology for Biomedical Investigations (OBI)
- Resource Description Framework (RDF)
- Schema.org
- Sleep Domain Ontology (SDO)
- Translational Data Catalog (IMI Data Catalog)
- UBER anatomy ONtology (UBERON)
- Uniform Resource Identifier (URI)
- Units Ontology (UO)
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
University of Luxembourg |
Writing - original draft |
||||
University of Oxford |
Writing - review & editing |